Figure 6.
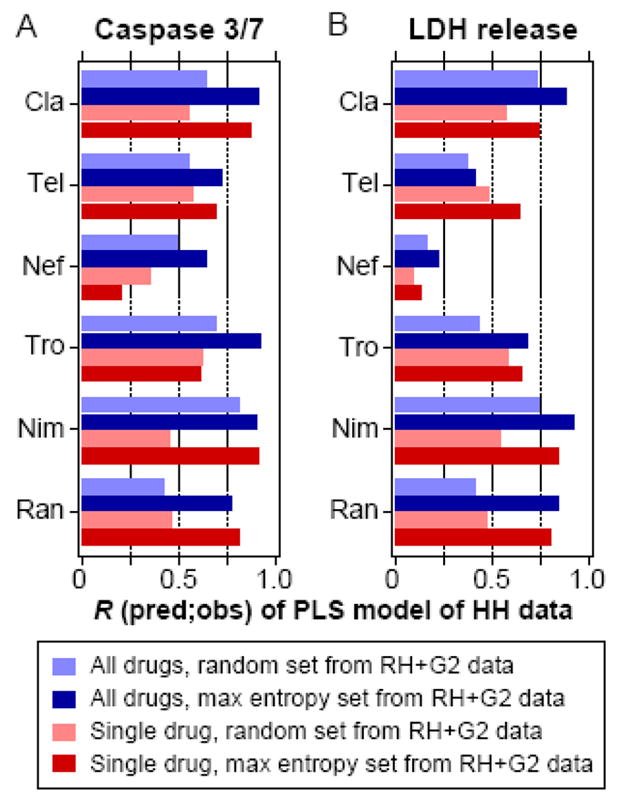
Predictive performance of partial least-squares (PLS) models trained on consensus maximum entropy sets and evaluated across cell systems. PLS regression models were built using specific cytokine condition sets containing 25 of the 32 possible cytokine treatments, selected based on data from rat hepatocytes (RH) and HepG2 cells (G2). They were evaluated for their ability to predict responses of the 7 remaining cytokine treatment conditions observed in human hepatocytes (HH). Pearson correlations (R) between the observed and predicted responses in HH of both the caspase 3/7 activity (panel A) and LDH release (panel B) responses are shown. Cytokine condition sets selected from RH and G2 data were chosen using either the consensus maximum entropy treatment sets or random treatment sets, of which the mean performance of 1000 random treatment sets is plotted, and these conditions sets were selected and evaluated using data from either all six drugs or single drugs. Abbreviations: Cla, clarithromycin; Tel, telithromycin; Nef, nefazodone; Tro, trovafloxacin; Nim, nimesulide; Ran, ranitidine.
