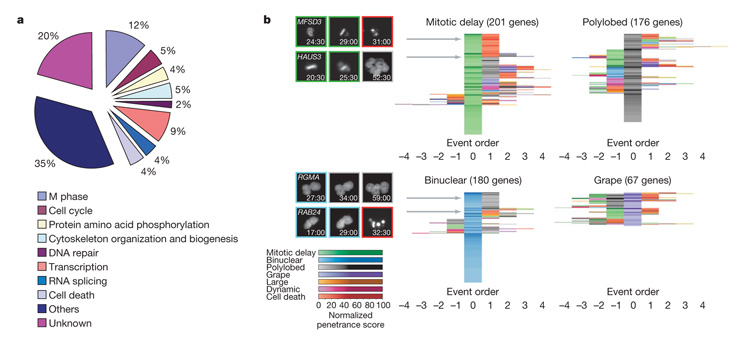
Figure 3. Gene Ontology (GO) terms distribution and event order maps.
a, GO analysis. Biological process annotations of the 572 validated mitotic hits. To deal with multiple annotations for each gene, categories were arbitrarily ordered by relevance to mitosis and each gene was assigned to the first term from this list that it had been annotated with. The same analysis was performed for the whole set of potential hit genes (1,249) in Supplementary Fig. 3. b, Event order maps. To each gene of the validated mitotic hit list (found by at least two siRNAs), a representative order of phenotypic events and a normalized penetrance score in each of the morphological classes is assigned (Supplementary Methods). For each gene, the event order can be visualized by a sequence of coloured fields where different colours correspond to different phenotypic classes, the colour intensity to the corresponding penetrance and the colour order to the phenotypic event order. The event orders are then centred on the phenotypic classes mitotic delay, polylobed, binuclear and grape. Genes with similar event orders are grouped together, resulting in centred event order maps, where the rows correspond to genes and the columns to the event order relative to the main phenotypic event. On the left, the locations of four genes (MFSD3, HAUS3, RMA, RAB24) in the maps are indicated by grey arrows; the corresponding order of phenotypic events are illustrated on a single cell level (numbers indicate time after transfection (h:min)). The corresponding event order maps for the whole set of potential hit genes (1,249) are shown in Supplementary Fig. 4. Single gene resolution event order maps are shown in Supplementary Fig. 5.