Table 2. Parameter combinations for the 10 best-fit simulations for the Great Lakes region as validated with Illinois P&I mortality data.
| Rank | RMS Error | Correlation Coefficient (r) | L (years) | D (days) |

|
 *
*
|
| 1 | 0.0049 | 0.93 | 5.74 | 4.58 | 5.11 | 2.35 |
| 2 | 0.0061 | 0.87 | 3.81 | 2.08 | 3.33 | 2.84 |
| 3 | 0.0062 | 0.84 | 9.78 | 2.68 | 4.92 | 1.90 |
| 4 | 0.0062 | 0.86 | 3.54 | 3.60 | 2.85 | 3.18 |
| 5 | 0.0064 | 0.86 | 3.66 | 3.29 | 3.67 | 2.29 |
| 6 | 0.0064 | 0.88 | 4.39 | 6.47 | 7.13 | 2.69 |
| 7 | 0.0065 | 0.83 | 7.65 | 2.42 | 2.79 | 3.69 |
| 8 | 0.0066 | 0.82 | 4.59 | 2.71 | 3.30 | 2.34 |
| 9 | 0.0069 | 0.82 | 4.81 | 6.53 | 3.15 | 3.32 |
| 10 | 0.0069 | 0.91 | 7.41 | 5.80 | 7.71 | 2.14 |
3000 simulations were performed at each site with the parameters
L (mean duration of immunity), D
(mean infectious period), ϕ (vitamin D scaling),
and R
0
* (the basic
reproduction number if
γi,t = 1) randomly
chosen from within specified ranges. Parameters λ
(inflection point) and η (inflection point slope)
were fixed at  and
and
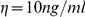 . Best-fit
simulations were selected based on RMS error after scaling the 31-year
mean daily infection number to the 31-year mean observed daily excess
P&I mortality rate.
. Best-fit
simulations were selected based on RMS error after scaling the 31-year
mean daily infection number to the 31-year mean observed daily excess
P&I mortality rate.
