Abstract
The mammalian totipotent and pluripotent lineage exhibits genome-wide dynamics in respect to DNA methylation content. The first phase of global DNA demethylation and de novo remethylation occurs during preimplantation development and gastrulation, respectively, while the second phase occurs in primordial germ cells and primary oocytes/prospermatogonia, respectively. These dynamics are indicative of a comprehensive epigenetic resetting or reprogramming of the genome in preparation for major differentiation events. To gain further insight into the mechanisms driving DNA methylation dynamics and other types of epigenetic modification, we performed an RNA expression microarray analysis of fetal prospermatogonia at the stage when they are undergoing rapid de novo DNA remethylation. We have identified a number of highly or specifically expressed genes which could be important for determining epigenetic change in prospermatogonia. These data provide a useful resource in the discovery of molecular pathways involved in epigenetic reprogramming in the mammalian germ line.
Keywords: epigenetics, DNA methylation, prospermatogonium, oogonium, pachytene spermatocyte, microarray
INTRODUCTION
In mammalian cells, the methylation of cytosines in CpG dinucleotides functions as an epigenetic modifier. DNA methylation regulates transcription of single copy genes, including imprinted genes and retrotransposons (Hojman-Montes de Oca et al., 1984; Feenstra et al., 1986; Hata and Sakaki, 1997; Szabo et al., 2000), and is required for maintaining genome stability—for example, chromosome segregation and X chromosome inactivation (Riggs and Singer-Sam, 1993). The large majority of DNA methylation is present at repetitive DNA, which comprises approximately one half of the genome. This DNA is represented by tandem head-tail repeats localised at centromeres, and by interspersed transposable elements (Hastie, 1989; Choo, 2001; Druker and Whitelaw, 2004).
The mammalian totipotent and pluripotent lineage—the zygote, cleavage-stage ovum, blastocyst inner cell mass, primitive ectoderm, and the germ line—behaves very differently to somatic cells in respect to DNA methylation. This is probably also true for other epigenetic modifiers, although these are less defined. While somatic cells do not change significantly in total methylation content at any stage, the totipotent and pluripotent lineage undergoes two phases of large-scale DNA de- and re-methylation during development and differentiation. In the first phase, the paternal genome is rapidly demethylated after the sperm nucleus enters the egg (Oswald et al., 2000). During cleavage, DNA methylation is lost probably passively so that the DNA of blastocysts is hypomethylated. Remethylation begins in the blastocyst and continues through gastrulation. By 7½ days post coitum (dpc) all lineages are fully remethylated (Monk et al., 1987). In the second phase, methylation is lost in primordial germ cells (PGCs). PGCs are derived from the primitive ectoderm (Lawson and Hage, 1994) at a stage when de novo methylation of DNA is still occurring, and the methylation status of the first PGCs to appear at 7½ dpc is unknown. Nevertheless, at 11½ dpc, soon after their arrival at the genital ridge, PGCs are globally hypomethylated (Monk et al., 1987), and imprinted gene sequences are hypomethylated by 13½ dpc (Brandeis et al., 1993; Hajkova et al., 2002; Lee et al., 2002). By 13½ dpc, female germ cells have entered meiosis (oogonia) but do not undergo significant DNA remethylation until much later during oocyte growth (Obata and Kono, 2002; Lucifero et al., 2004). By contrast, at 13½ dpc, male germ cells are entering mitotic arrest (prospermatogonia) with de novo DNA methylation commencing shortly after and being complete by birth. Sequences that become methylated at this stage include retrotransposons (Monk et al., 1987) and imprinting control regions (Davis et al., 2000; Ueda et al., 2000).
Each period of global DNA methylation erasure occurs immediately before a major phase of differentiation and development—erasure during pre-implantation development is followed by gastrulation, while erasure in PGCs is followed by sexual differentiation and the beginning of gamete differentiation. These juxtapositions are probably not accidental and are indicative of a comprehensive re-setting of the epigenome in preparation for major differentiation events. If so, then the regulatory components determining DNA methylation dynamics are therefore likely to be a component of the overall mechanism of epigenetic de- and re-programming in the totipotent and pluripotent lineage. Learning more of germ cell DNA methylation dynamics should therefore help in understanding the epigenetic programming essential for successful embryonic development, germ cell development, and reproduction.
To identify proteins that could be involved in regulating DNA methylation dynamics and other forms of epigenetic modification in prospermatogonia, we performed an RNA expression microarray analysis in purified cells at 15½ dpc. This is the stage when prospermatogonia are undergoing rapid de novo DNA methylation. For comparison, we also analysed 15½ dpc pachytene oogonia, 15½ dpc female and male gonadal somatic cells, and adult pachytene spermatocytes. Here we describe a number of genes encoding epigenetic modifiers that are expressed at relatively high levels in prospermatogonia, and which are candidates for regulating epigenetic modifications during their development.
RESULTS AND DISCUSSION
The microarray platform used was the GeneChip Mouse Expression Set 430, consisting of two chips, 430A and 430B (Affymetrix). This platform provides a combined coverage of 45,000 probe sets to analyse the expression level of over 39,000 transcripts and variants of over 34,000 well characterised mouse genes. All raw data obtained has been deposited in the EMBL-EBI ArrayExpress database: Accession number E-TABM-412.
Expression of germ cell markers
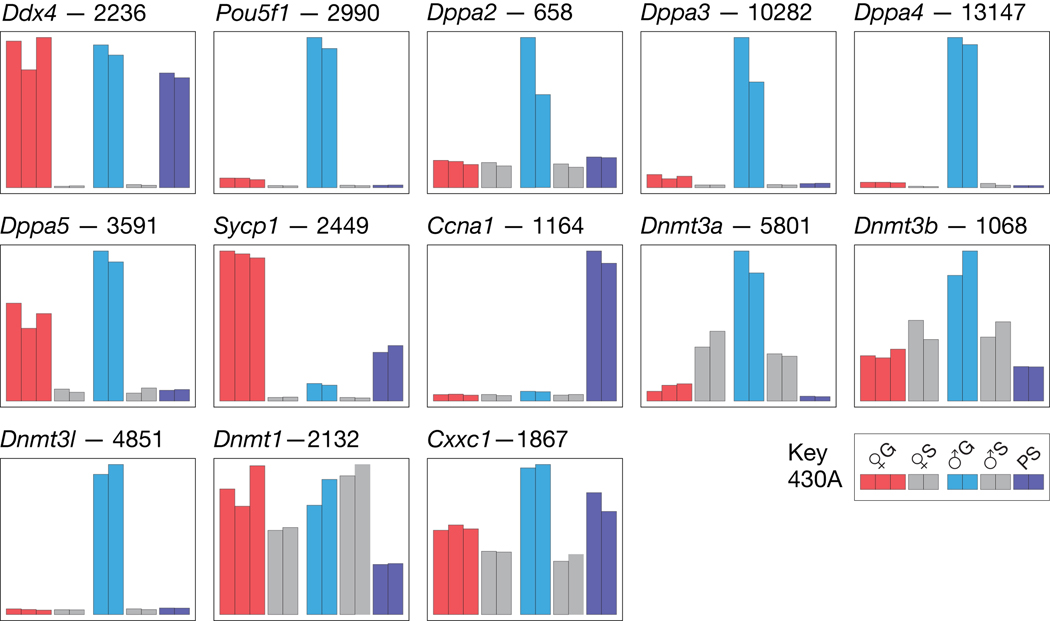
In all cases examined, the relative expression of germ cell markers in the various samples conformed to expectation. This demonstrates the high purity and quality of the RNA samples, and provides assurance that the data set is an accurate read out of gene specific expression in the cell types collected. Relative expression of selected markers is shown (Fig. 1). General germ cell markers are DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 (Ddx4) (Fujiwara et al., 1994), POU domain, class 5, transcription factor 1 (Pou5f1) (Yeom et al., 1996), and developmental pluripotency associated 2, 3, 4 and 5 (Dppa2, Dppa3, Dppa4 and Dppa5) (Bowles et al., 2003; Western et al., 2005; Maldonado-Saldivia et al., 2007). As expected, all show specific expression in 15½ dpc germ cells. The significantly greater amount of Pou5f1 RNA in prospermatogonia relative to oogonia is reflected by a Pou5f1-gfp transgene (Szabo et al., 2002). Also as expected, synaptonemal complex protein 1 (Sycp1) RNA is specific to pachytene oogonia and spermatocytes (Sage et al., 1999), while cyclin A1 (Ccna1) RNA is expressed highly only in pachytene spermatocytes (Liao et al., 2004) (Fig. 1). Relative expression of selected genes was validated by quantative real-time PCR (QPCR) analysis, discussed below.
Fig. 1.
Relative expression of germ cell marker genes. At the top left of each histogram is given the gene symbol followed by the intensity value for the sample showing the highest relative level of expression. Key: ♀G, 15½ dpc female germ cells (oogonia, at meiosis I, pachytene); ♀S, 15½ dpc female gonadal somatic cells; ♂G, 15½ dpc male germ cells (prospermatogonia, in mitotic arrest); ♂S, 15½ dpc male gonadal somatic cells; PS, adult spermatocytes (at meiosis I, pachytene). All 13 genes are on the 430A chip. Probes sets corresponding to the histograms: Ddx4, 1427242_at; Pou5f1, 1417945_at; Dppa2, 1429654_at; Dppa3, 1424295_at; Dppa4, 1429597_at; Dppa5, 1416552_at; Sycp1, 1427291_at; Ccna1, 1449177_at; Dnmt3a, 1423066_at; Dnmt3b, 1418351_a_at; Dnmt3l, 1425035_s_at; Dnmt1, 1435122_x_at; Cxxc1, 1452221_a_at. When the expression of a gene is represented by multiple probe sets, only the probe set giving the highest intensity values was used to generate the histogram.
General comparison of gene expression in germ and somatic cells
Results from the 430A and 430B chips were combined. Statistical contrast analysis for expression in prospermatogonia, oogonia, male and female gonadal somatic cells was applied using the false discovery rate adjustment method (FDR) and the more stringent HOLM method. Numbers of genes falling into expression level categories are shown in Table 1.
TABLE 1.
Number of genes preferentially expressed in prospermatogonia and oogonia
| Statistical contrast analysis method | ||
|---|---|---|
| n-fold* | FDR | HOLM |
| Prospermatogonia and oogonia combined | ||
| all | 2162 | 130 |
| 2 | 1061 | 122 |
| 3 | 391 | 110 |
| 5 | 129 | 79 |
| 10 | 46 | 43 |
| Prospermatogonia | ||
| all | 2266 | 114 |
| 2 | 983 | 112 |
| 3 | 330 | 94 |
| 5 | 101 | 58 |
| 10 | 28 | 20 |
| Oogonia | ||
| all | 3199 | 471 |
| 2 | 1410 | 454 |
| 3 | 513 | 332 |
| 5 | 140 | 129 |
| 10 | 33 | 32 |
n-fold higher expression relative to all remaining cell types combined
Prospermatogonia and oogonia combined versus male and female somatic cells combined: The gene set showing over 5-fold higher expression in germ cells is moderately enriched in genes associated with germ cell development, meiosis, and the meiotic cell cycle (5-fold set: 129 genes, Table 1; Supplementary Data—List 1) (P-adjusted < 0.01 using FuncAssociate: <http://llama.med.harvard.edu/cgi/func/funcassociate>). In the gene set with 3-fold higher expression in germ cells, approximately 20% of the probe sets represent poorly annotated transcripts.
Prospermatogonia versus oogonia, male and female somatic cells combined: Approximately 18% of probe sets detected at 3-fold higher expression in prospermatogonia correspond to poorly annotated transcripts (3-fold set: 330 genes, Table 1; Supplementary Data—List 2). The genes Dppa3 and Dppa4 are among the most highly differentially expressed genes in prospermatogonia, while Dppa2 expression is also specific to prospermatogonia although in lower amounts. DNA (cytosine-5-)-methyltransferase 3-like (Dnmt3l), DNA methyltransferase 3A (Dnmt3a), piwi-like homologue 2 (Drosophila) (Piwil2), and piwi-like homologue 4 (Drosophila) (Piwil4) are present in this prospermatogonia-specific set, and are discussed further below.
Oogonia versus prospermatogonia and male and female somatic cells combined: Relative to prospermatogonia, a larger number of genes are preferentially expressed in oogonia (3-fold set: 513 genes, Table 1; Supplementary Data—List 3). In this 3-fold set, approximately 25% of probe sets represent poorly annotated transcripts. Fragile X mental retardation 1 neighbour (Fmr1nb) was one of the most highly expressed genes in this 3-fold set, which also contained DNA methyltransferase 2/tRNA aspartic acid methyltransferase 1 (Dnmt2/Trdmt1). Genes expressed specifically in oogonia were strongly associated with meiosis, DNA metabolism and chromosome assembly or pairing (P > 0.001).
Expression of epigenetic modifiers in relation to DNA methylation
DNA methyltransferase (DNMT) enzymes are required for de novo DNA methylation in germ cells—in particular, DNMT3A, for which activity in greatly enhanced by DNMT3L (Bourc’his et al., 2001; Hata et al., 2002; Lehnertz et al., 2003; Bourc’his and Bestor, 2004; Kaneda et al., 2004; Chen et al., 2005; Webster et al., 2005). The expression of genes encoding these two DNMTs, and Dnmt3b, are high in prospermatogonia, with the expression of Dnmt3l being essentially specific as described for prospermatogonia at 2 days post partum (Bourc’his and Bestor, 2004) (Fig. 1). DNMT1, best known as the predominant maintenance DNA methyltransferase, is expressed appreciably in all cell types (Fig. 1) and could play some role in de novo methylation in prospermatogonia. Further, the CCXC finger 1 (PHD domain) gene (Cxxc1) is expressed most strongly in prospermatogonia (Fig. 1). The CXXC1 protein is required for global DNA methylation content in mouse embryonic stem cells, and in this function possibly acts in concert with DNMT1 (Carlone et al., 2005).
DNMTs do not act alone in methylating DNA. Evidence is mounting that DNA methylation and histone post-translational modifications are directly connected. For example, (1) in plants and fungi, histone H3 lysine 9 (H3K9) di- and tri-methylation, respectively is a prerequisite for DNA methylation (Tamaru et al., 2003; Jackson et al., 2004), (2) in higher organisms, RNA interference (RNAi) mechanisms direct the establishment of H3K9 tri-methylation at centromere repeats, which in turn directs heterochromatinsation and DNA methylation (Lehnertz et al., 2003), (3) H3K27 methylation is enriched at silenced homoeotic genes, the inactive X-chromosome, and imprinted genes (Martin and Zhang, 2005), and therefore may play a role in the recruitment of DNA methylation, (3) changes in the content of certain histone modifications are observed in DNMT3L deficient spermatogenic cells (Webster et al., 2005). It is probably also likely that active or passive DNA demethylation is also linked with underlying patterns of histone post-translational modifications. For example, DNA methylation at imprinting control regions of imprinted genes is resistant to demethylation during preimplantation development (Olek and Walter, 1997; Warnecke and Clark, 1999; Nakamura et al., 2007). This could be explained by a unique histone modification that can attract maintenance DNA methyltransferase activity.
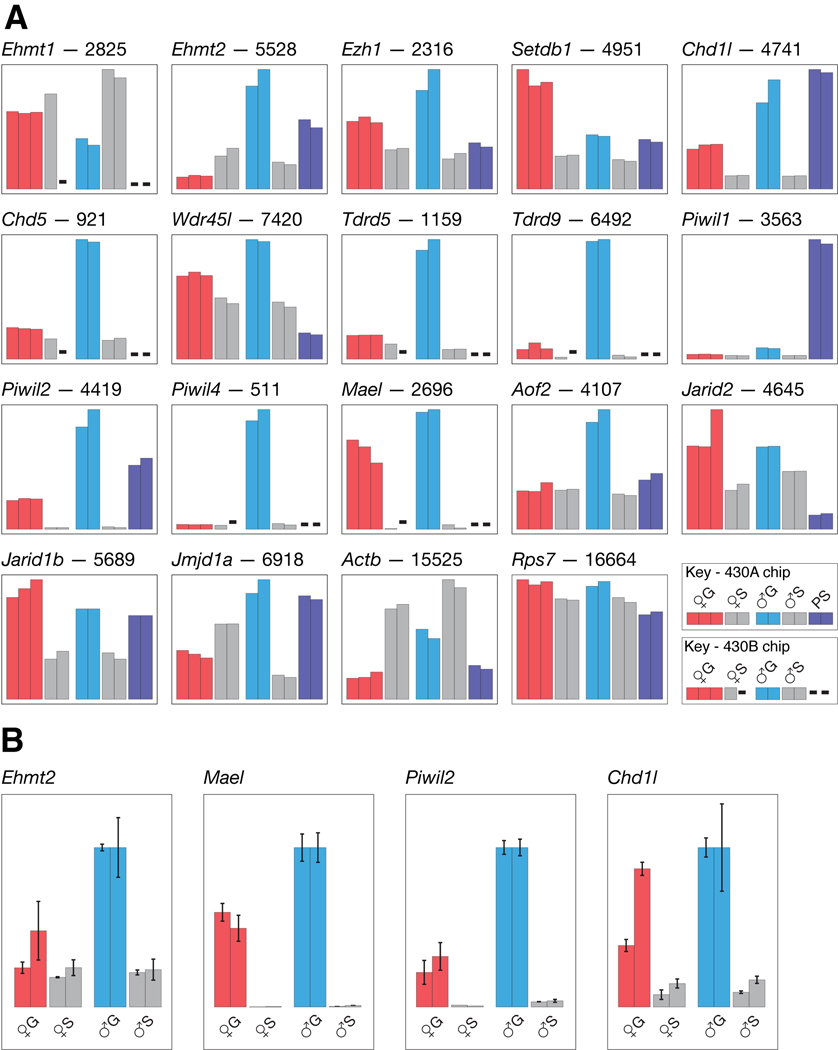
Enzymes that carry out methylation of H3K9 and H3K27 belong to a family of proteins classified according to the presence of a conserved ~125 aa SET domain. SET domain enzymes known to methylate H3K9 and H3K27 (Dillon et al., 2005; Martin and Zhang, 2005). Suppressor of variegation 3–9 homologue 1 (SUV39H1) and suppressor of variegation 3–9 homologue 2 (SUV39H2) together are essential for normal heterochromatin formation at peri-centric repeats in male germ cells (Peters et al., 2001). Neither Suv39h1 or Suv39h2 appears to be expressed at a high level in prospermatogonia (data not shown). Higher expression may be required only at later stages, when Suv39h1-Suv39h2 double-knockout mice exhibit abnormalities and deficiencies in centromere DNA methylation in meiosis (Peters et al., 2001). Global H3K9 and H3K27 methylation is carried out by euchromatic histone methyltransferase 1 (EHMT1) and euchromatic histone lysine N-methyltransferase 2 (EHMT2). While they are present in the same complex, their functions are not redundant, at least in embryonic stem cells (Tachibana et al., 2005). DNMT1 and EHMT2 interact in the coordination of DNA and histone methylation during replication (Esteve et al., 2006), and EHMT2 is also required for de novo DNA methylation of the Pou5f1 promoter by DNMT3A or DNMT3B in embryonic stem cell differentiation (Feldman et al., 2006). The finding that EHMT2 can target DNA methylation to specific sequences raises the possibility that EHMT2-mediated H3K9 methylation may perform a similar function in prospermatogonia—for example, the targeting of DNA methylation to retrotransposon promoters and to regions of imprinted genes, such as the paternally methylated imprinting control region of the insulin-like growth factor 2 and H19 genes (Mann et al., 2000). Consistent with this possibility, Ehmt2 RNA is abundant in prospermatogonia (Fig. 2A and B) while the RNA concentration of Ehmt1 is appreciable (Fig. 2A). At least two other genes encoding for SET domain proteins exhibit high expression in prospermatogonia, and are thereby implicated in the establishment of DNA methylation and other forms of epigenetic modification. These are enhancer of zeste homologue (Drosophila) (Ezh1), and SET domain, bifurcated 1 (Setdb1) (Fig. 2A).
Fig. 2.
Relative expression of selected genes. A: Microarray. Thirteen and six genes on the 430A and 430B chips, respectively, are shown. Probes sets corresponding to the histograms: Ehmt1, 1454776_at; Ehmt2, 1460692_at; Ezh1, 1449023_a_at; Setdb1, 1451833_a_at; Chd1l, 1449415_at; Chd5, 1436095_at; Wdr45l, 1416281_at; Tdrd5, 1456391_at; Tdrd9, 1453357_at; Piwil1, 1460340_at; Piwil2, 1449170_at; Piwil4, 1447564_x_at; Mael, 1436837_at; Aof2, 1426762_s_at; Jarid2, 1422698_s_at; Jarid1b, 1427143_at; Jmjd1a, 1426810_at; Actb, 1436722_a_at; Rps7, 1419364_a_at. Other details are as given in the legend to Figure 1. B: Quantitative PCR. Two independent reverse transcription and QPCR assays, represented by the bar pairs, were performed on four selected transcripts. Each bar is the mean of triplicate reactions, with the total size of the error bar equal to twice the standard deviation. Expression relative to prospermatogonia (♂G) is on a linear scale, with Rps7 expression used for normalisation. Normalising to Actb expression results in the highest relative values being present in oogonia (data not shown). This nomalisation is almost certainly spurious as the amount of Actb transcript in oogonia is considerably lower than in the other three cell types as indicated in the microarray analysis (Fig. 2A) and QPCR assays (data not shown).
After H3K9 and H3K27 have been methylated, these post-translational modifications are recognised by proteins that perform downstream functions. To date, at least three protein motifs have been identified that can interact with histone lysine modifications—(1) the chromodomain of CBX5 (HP1), which interacts with methylated H3K9 in the formation of constitutive heterochromatin at centromeres, and the chromodomain of CBX4 contained in the polycomb repressive complex 1, which interacts with methylated H3K27 in the formation of faculative heterochromatin in, for example, the inactive X chromosome. These proteins are therefore required in the formation of repressive chromatin conformations and possibly recruit DNMTs, (2) the WD-repeat domain contained in WDR5 binds to di-methylated H3K4 and is needed for its conversion to the tri-methylated state. This modification is involved in transcriptional activation (Dillon et al., 2005; Martin and Zhang, 2005), and (3) the Tudor domain, which binds methylated H3K79 and is involved in DNA repair (Dillon et al., 2005; Martin and Zhang, 2005). In mammals, for each of these three conserved domains there exists a number of protein paralogues. Some of these exhibit high, and often also specific, expression in prospermatogonia, thereby implicating functional significance of encoded proteins as epigenetic modifiers. Examples shown are chromodomain helicase DNA binding protein 1-like (Chd1l) (Fig. 2A and B), chromodomain helicase DNA binding protein 5 (Chd5), Wdr45-like (Wdr45l), tudor domain containing 5 (Tdrd5), and tudor domain containing 9 (Tdrd9) (Fig. 2A).
Histone demethylation could also play a role in modifying the epigenetic profile of prospermatogonia. The transcript encoding amine oxidase (flavin containing) domain 2 (AOF2), a H3K4 demethylase (Shi et al., 2004), is high in prospermatogonia (Fig. 2A), as are transcripts encoding some of the jumonji domain containing proteins which function as histone demethylases. These are jumonji, AT rich interative domain 2 (Jarid2), jumonji, AT rich interative domain 1B (Rbp2 like) (Jarid1b) and jumonji domain containing 1A (Jmjd1a) (Fig 2A). JMJD1A is a H3K9 demethylase (Yamane et al., 2006) and conceivably could fine tune EHMT2-mediated H3K9 methylation patterns. Interestingly, JMJD1A deficiency is associated with a failure of spermiogenesis, or post-meiotic male germ cell development. Whether DNA methylation is affected in these mutants is unclear (Okada et al., 2007). Most of genes encoding jumonji domain containing proteins are poorly expressed in prospermatogonia (data not shown).
In mammalian cells, RNAi is involved in the silencing of retrotransposons (Yang and Kazazian, 2006), and it can be speculated that this is accomplished through histone post-translational modification intermediates in a similar way as is achieved for centomeres and peri-centric regions. In addition, small RNAi-independent non-coding RNAs that have been found only in spermatogenic cells—piwi-like homologue (Drosophila) (PIWIL)-associated RNAs (piRNAs) (Grivna et al., 2006; Watanabe et al., 2006)—are complexed with proteins that are essential for spermatogenesis and required for the presence of nomal amounts of DNA methylation in spermatogenic cells. These proteins are PIWIL2 (Kuramochi-Miyagawa et al., 2004; Aravin et al., 2007) and PIWIL4 (Carmell et al., 2007). Piwil2 RNA is abundant in prospermatogonia (Fig. 2A and B), while Piwil4 RNA is specific to these cells (Fig. 2A). Further, the expression of other genes for which the encoded proteins are present in PIWIL protein complexes are high in prospermatogonia. These are Ddx4 (Fig. 1) and maelstrom homologue (Drosophila) (Mael) (Costa et al., 2006) (Fig. 2A and B). These findings raise the possibility that PIWIL and associated proteins are involved in de novo DNA methylation in prospermatogonia. It is conceivable that the steps involve the targeting of stretches of CpGs by piRNAs in PIWIL/EHMT complexes, followed by histone modification-directed DNA methylation. It is also possible that DNA methyltransferases are complexed with PIWILs and do not depend on histone modifications for DNA methylation.
These microarray data provide a useful resource to assist in the discovery of molecular pathways that determine de novo DNA remethylation and other epigenetic modifications in mammalian prospermatogonia, and that are components of the general mechanism of epigenetic reprogramming essential for germ cell development. These data provide an excellent starting point for further analysis through mutagenesis strategies, and are also relevant for other studies which would benefit from a comprehensive read-out of gene expression at the stages of germ cell development analysed here.
EXPERIMENTAL PROCEDURES
Purification of germ cells
Fetal germ cells and gonadal somatic cells: To obtain 15½ dpc fetuses for germ cell isolation, outbred CD-1 adult females (Charles River Laboratories) were mated to males homozygous for a transgene in which the green fluorescent protein (gfp) reporter coding sequence is driven by regulatory sequences of the Pou5f1 gene. In this line, GFP is expressed specifically in germ cells (Szabo et al., 2002) (Jackson Laboratory stock no. 004654). Fetuses were dissected free of the uterus, immediately decapitated, then placed in phosphate buffered saline on ice. Up to 10 isolated testes or ovaries were dissected free of the metanephros, incubated in 0.15 mL 0.25% trypsin-EDTA (Gibco) in medium M2 (Wood et al., 1987) (37°C, 20 min) in a 1.5 mL centrifuge tube, triturated with a 0.2 mL tip to obtain a single cell suspension, 0.3 mL of medium M2 containing 10% fetal bovine serum added, the cell suspension passed through 70 µM mesh, then placed on ice. GFP-positive germ cells and GFP-negative gonadal somatic cells were sorted in the same run using a MoFlo MLS flow cytometer (Dako). Testes isolated from four pregnant females yielded approximately 2 × 105 sorted germ cells, which constituted one sample, and likewise for ovaries. One million testicular somatic cells were sorted, which constituted one sample for RNA purification, and likewise for ovarian somatic cells. Sorted cells were pelleted, then total RNA isolated using an RNeasy kit (Qiagen).
Adult pachytene spermatocytes: From one male, isolated testes were dissected free of the capsules, teased apart in DPBSGB (Dulbecco’s phosphate buffered saline with calcium and magnesium supplemented with 5.56 mM glucose and 2 mg/mL BSA), transferred to a 50 mL centrifuge tube containing 5 mL of 0.2 mg/mL collagenase type 1 (Worthington Biochemical Corp.) in DPBSGB, agitated (37°C, 5 min), dispersed by pipetting with a 5 mL pipette, then agitated again (37°C, 5 min). Tubules were allowed to settle by gravity, then the supernatant with interstitial cells removed. Tubules were washed with 5 mL DPBSGB by swirling, allowed to settle again, then the supernatant removed. 5 mL of 0.25% trypsin-EDTA and 5 µL of 10 U/mL DNase I, RNase-free (Roche Applied Science) were added to the tube and the tubules incubated with occasional agitation (37°C, 15 min). 1 mL of fetal bovine serum was added, then the tubules converted to a cell suspension by pipetting with a 5 mL pipette. The cell suspension was passed through a 70 µM mesh cell strainer (Falcon) into a new 50 mL centrifuge, cells pelleted (250 × g, 5 min), then the pellet resuspended in 5 mL of DPBSGB containing 5% fetal bovine serum. The suspension was mixed with 5 µL of 10 mg/mL Hoechst 33342 (Molecular Probes), incubated (37°C, 20 min), then placed on ice. Immediately before flow cytometry, the suspension was mixed with 5 µL of 1 mg/mL propidium iodide (Molecular Probes). Pachytene spermatocytes were sorted on the basis of (1) 4c genome content—the most fluorescent of three well separated peaks on the Hoechst scale, (2) larger size—events with higher forward and side scatter properties, and (3) viability—the population with the lowest level of propidium iodide staining. Examination by microscopy of sorted cells indicated a high purity of pachytene spermatocytes on the basis of morphology. One million cells were sorted, which constituted one sample for RNA purification.
Microarray analysis
Between 0.5 and 1.0 µg of total RNA was obtained from each sample, which was amplified once before hybridisation to chips. The data was normalised in Bioconductor (Gentleman et al., 2004) using the RMA function of the Affy package (Irizarry et al., 2003). Quality was assessed by clustering, outliers were discarded, and statistical analysis of differential expression was performed with the limma package (Smyth, 2004).
Quantitative assessment of relative RNA concentrations
The relative amounts or RNA in female and male germ and somatic cells was determined by QPCR. Germ and somatic cells of 15½ dpc gonads were purified as described above. RNA was purified using the mirVana miRNA Isolation Kit (Ambion) and contaminating DNA removed with the DNA-free Kit (Ambion). Reverse transcription (RT) was carried out in 20 µL reaction volumes using 100 ng RNA, 1 mM final concentration of total dNTPs (Bioline), 50 µM final concentration of Random Primer 12 (New England BioLabs) and M-MuLV reverse transcriptase and reaction buffer (New England BioLabs). RT reaction conditions were 64°C-10 m → 25°C-10 m, with reverse transcriptase added at 8–10 m → 42°C-30 m → 99°C-5 m. Gene transcripts analysed and pre-validated primer pairs (identification numbers) used in QPCR reactions were: Ehmt2, 22219432a1; Mael, 30424966a1; Piwil2, 10946610a1; Chd1l, 13386044a1 (Primer Bank <http://pga.mgh.harvard.edu/primerbank/>) (Wang and Seed, 2003). Primers were further assessed and validated by standard curve analysis. For each transcript, amplicons correspond to a region well upstream of that corresponding to the Affymetrix probe set (Fig. 2). Transcripts for normalisation and primers: Actin, beta, cytoplasmic (Actb) (Carmell et al., 2007), and ribosomal protein S7 (Rps7), upper 5′-TGG TCT TCA TTG CTC AGA GGA GG-3′, lower 5′-TGC CAT CCA GTT TCA CAC GG-3′. Reactions, in triplicate, were carried out using ABsolute Blue QPCR SYBR Green Mix (ABgene) and 100 nM of each primer in 20 µL. Reactions and analysis was carried out using the Mx3000P QPCR System (Stratagene).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Analytical Cytometry Core of the City of Hope for assistance in purifying germ cells, Piroska Szabo for purifying RNA, and the University of California at Irvine DNA and Protein Microarray Facility for performing the microarray assays.
Grant sponsor: NHMRC, grant numbers: 350216 and 350217
Grant sponsor: NIH, grant number: R01-GM64378
REFERENCES
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking DNMT3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. DNMT3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Bowles J, Teasdale RP, James K, Koopman P. DPPA3 is a marker of pluripotency and has a human homologue that is expressed in germ cell tumours. Cytogenet Genome Res. 2003;101:261–265. doi: 10.1159/000074346. [DOI] [PubMed] [Google Scholar]
- Brandeis M, Kafri T, Ariel M, Chaillet JR, McCarrey J, Razin A, Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlone DL, Lee JH, Young SR, Dobrota E, Butler JS, Ruiz J, Skalnik DG. Reduced genomic cytosine methylation and defective cellular differentiation in embryonic stem cells lacking CpG binding protein. Mol Cell Biol. 2005;25:4881–4891. doi: 10.1128/MCB.25.12.4881-4891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Mann JR, Hsieh CL, Riggs AD, Chedin F. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J Cell Biochem. 2005;95:902–917. doi: 10.1002/jcb.20447. [DOI] [PubMed] [Google Scholar]
- Choo KH. Domain organization at the centromere and neocentromere. Dev Cell. 2001;1:165–177. doi: 10.1016/s1534-5807(01)00028-4. [DOI] [PubMed] [Google Scholar]
- Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. Mouse MAELSTROM: The link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet. 2006;15:2324–2334. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druker R, Whitelaw E. Retrotransposon-derived elements in the mammalian genome: a potential source of disease. J Inherit Metab Dis. 2004;27:319–330. doi: 10.1023/B:BOLI.0000031096.81518.66. [DOI] [PubMed] [Google Scholar]
- Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9A coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra A, Fewell J, Lueders K, Kuff E. In vitro methylation inhibits the promotor activity of a cloned intracisternal A-particle LTR. Nucleic Acids Res. 1986;14:4343–4352. doi: 10.1093/nar/14.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9A-mediated irreversible epigenetic inactivation of OCT-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Komiya T, Kawabata H, Sato M, Fujimoto H, Furusawa M, Noce T. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc Natl Acad Sci USA. 1994;91:12258–12262. doi: 10.1073/pnas.91.25.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Hastie ND. Highly repeated DNA families in the genome of Mus Musculus. In: Lyon MF, Searle AG, editors. Genetic Variants and Strains of the Laboratory Mouse. Oxford, New York, Tokyo: Oxford University Press; 1989. pp. 559–573. [Google Scholar]
- Hata K, Okano M, Lei H, Li E. DNMT3L cooperates with the DNMT3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- Hata K, Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- Hojman-Montes de Oca F, Lasneret J, Dianoux L, Canivet M, Ravicovitch-Ravier R, Peries J. Regulation of intracisternal A particles in mouse teratocarcinoma cells: Involvement of DNA methylation in transcriptional control. Biol Cell. 1984;52:199–204. doi: 10.1111/j.1768-322x.1985.tb00337.x. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Gautier L, Cope LM. An R package for analyses of Affymetrix oligonucleotide arrays. In: Parmigiani G, Garett ES, Irizarry RA, Zeger SL, editors. The Analysis of Gene Expression Data: Methods and Software. New York: Springer-Verlag; 2003. pp. 102–119. [Google Scholar]
- Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase DNMT3A in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of Piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Hage WJ. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp. 1994;182:68–84. doi: 10.1002/9780470514573.ch5. discussion 84–91. [DOI] [PubMed] [Google Scholar]
- Lee J, Inoue K, Ono R, Ogonuki N, Kohda T, Kaneko-Ishino T, Ogura A, Ishino F. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129:1807–1817. doi: 10.1242/dev.129.8.1807. [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH. SUV39H-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- Liao C, Li SQ, Wang X, Muhlrad S, Bjartell A, Wolgemuth DJ. Elevated levels and distinct patterns of expression of A-type cyclins and their associated cyclin-dependent kinases in male germ cell tumors. Int J Cancer. 2004;108:654–664. doi: 10.1002/ijc.11573. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13:839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- Maldonado-Saldivia J, van den Bergen J, Krouskos M, Gilchrist M, Lee C, Li R, Sinclair AH, Surani MA, Western PS. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2007;25:19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- Mann JR, Szabo PE, Reed MR, Singer-Sam J. Methylated DNA sequences in genomic imprinting. Crit Rev Eukaryot Gene Expr. 2000;10:241–257. doi: 10.1615/critreveukargeneexpr.v10.i3-4.30. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T. PGC7/STELLA protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Obata Y, Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J Biol Chem. 2002;277:5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- Olek A, Walter J. The pre-implantation ontogeny of the H19 methylation imprint. Nat Genet. 1997;17:275–276. doi: 10.1038/ng1197-275. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the SUV39H histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Riggs AD, Singer-Sam J. X chromosome inactivation and DNA methylation. In: Jost JP, Saluz HP, editors. DNA Methylation: Molecular Biology and Biological Significance. Basel: Birkhauser Verlag; 1993. pp. 358–384. [Google Scholar]
- Sage J, Martin L, Meuwissen R, Heyting C, Cuzin F, Rassoulzadegan M. Temporal and spatial control of the Sycp1 gene transcription in the mouse meiosis: regulatory elements active in the male are not sufficient for expression in the female gonad. Mech Dev. 1999;80:29–39. doi: 10.1016/s0925-4773(98)00191-9. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Szabo P, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol. 2000;10:607–610. doi: 10.1016/s0960-9822(00)00489-9. [DOI] [PubMed] [Google Scholar]
- Szabo PE, Hubner K, Scholer H, Mann JR. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev. 2002;115:157–160. doi: 10.1016/s0925-4773(02)00087-4. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9A and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Ueda T, Abe K, Miura A, Yuzuriha M, Zubair M, Noguchi M, Niwa K, Kawase Y, Kono T, Matsuda Y, Fujimoto H, Shibata H, Hayashizaki Y, Sasaki H. The paternal methylation imprint of the mouse H19 locus is acquired in the gonocyte stage during foetal testis development. Genes Cells. 2000;5:649–659. doi: 10.1046/j.1365-2443.2000.00351.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke PM, Clark SJ. DNA methylation profile of the mouse skeletal alpha-actin promoter during development and differentiation. Mol Cell Biol. 1999;19:164–172. doi: 10.1128/mcb.19.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: Retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster KE, O’Bryan MK, Fletcher S, Crewther PE, Aapola U, Craig J, Harrison DK, Aung H, Phutikanit N, Lyle R, Meachem SJ, Antonarakis SE, de Kretser DM, Hedger MP, Peterson P, Carroll BJ, Scott HS. Meiotic and epigenetic defects in Dnmt3l-knockout mouse spermatogenesis. Proc Natl Acad Sci USA. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western P, Maldonado-Saldivia J, van den Bergen J, Hajkova P, Saitou M, Barton S, Surani MA. Analysis of Esg1 expression in pluripotent cells and the germline reveals similarities with Oct4 and Sox2 and differences between human pluripotent cell lines. Stem Cells. 2005;23:1436–1442. doi: 10.1634/stemcells.2005-0146. [DOI] [PubMed] [Google Scholar]
- Wood MJ, Whittingham DG, Rall WF. The low temperature preservation of mouse oocytes and embryos. In: Monk M, editor. Mammalian Development: A Practical Approach. Oxford: IRL Press; 1987. pp. 255–280. [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yang N, Kazazian HHJ. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.