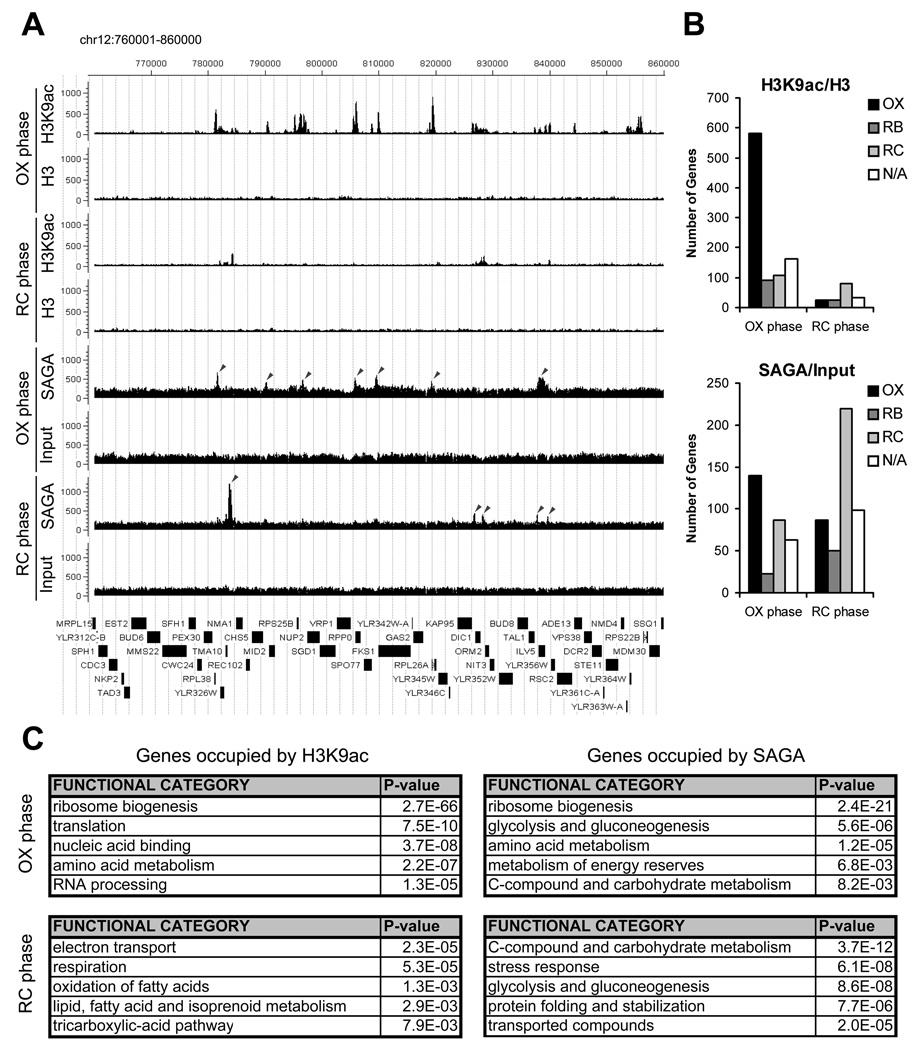
Figure 5. Genome-wide Analysis Reveals that Histone H3 at Growth Genes becomes Acetylated upon Entry into Growth.
(A) ChIP-seq analysis was performed to reveal genomic regions occupied by H3K9ac and SAGA at a time point in either early OX or early RC phase, as in Figure 4. Sequencing data were analyzed and displayed using CisGenome (Ji et al., 2008). The arrows mark peaks associated with the following genes (maximal YMC phase of expression in parentheses): SAGA OX phase - RPL38 (OX), MID2 (OX), RPS25B (OX), RPP0 (OX), FKS1 (OX), RPL26A (OX), ILV5 (OX); SAGA RC phase - TMA10 (RC), KAP95 (OX), DIC1 (RC), TAL1 (N/A), ILV5 (OX). The temporal expression profiles of these genes across the YMC were highly consistent with these ChIP-seq data (Figure S4). Note that H3K9ac peaks are much more frequent and significant in OX phase compared to RC phase. Many SAGA binding sites in OX phase corresponded directly to significant regions of H3K9 acetylation, while many SAGA binding sites in RC phase did not.
(B) YMC phase distribution of genes occupied by H3K9 acetylation or SAGA in either early OX or RC phase. Many more genomic regions of H3K9 acetylation were present in OX phase (n=942) compared to RC phase (n=161) at the same level of statistical significance. H3K9 acetylation was present almost exclusively at OX phase growth genes, specifically during OX phase (p-value < 9.8 × 10−140). As determined previously by microarray analysis, there are ~1,016 OX, 975 RB, and 1,508 RC phase genes in the YMC (Tu et al., 2005).
(C) Functional categories of genes occupied by H3K9 acetylation or SAGA as determined by MIPS (Munich Information Center for Protein Sequences) functional classification. The genes called by each ChIP-seq experiment were used to identify overrepresented gene functions (the top 100 genes are listed in Tables S1–S4). Note that genes involved in ribosome biogenesis were found to be the most significant group of H3K9ac and SAGA targets. Genes involved in translation, amino acid metabolism, and rRNA processing which are also critical for growth, are also highly overrepresented.