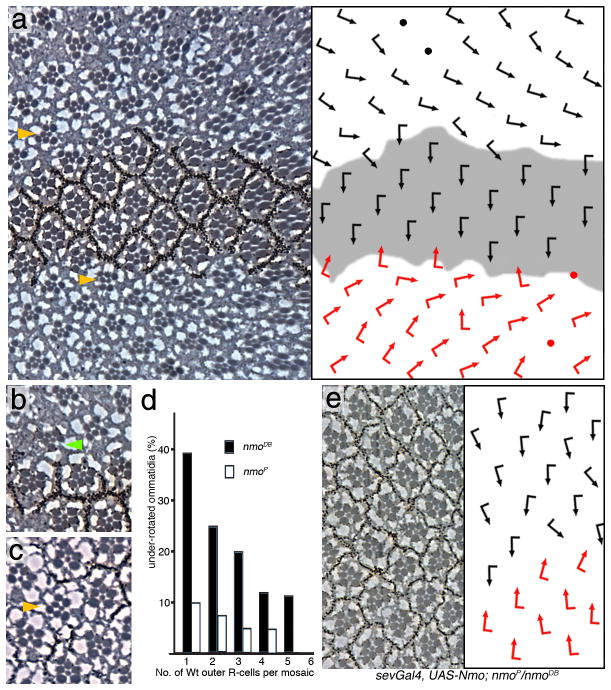
Figure 2. nmo is required in a cluster autonomous manner for rotation.
Panels a–c, and e show tangential sections of adult eyes, (d) shows a quantification; dorsal is up and anterior left. The respective schematic presentations of ommatidial orientation (arrows as in Fig. 1) are shown as right panels in a and e. (a–c) Mutant clones of nmoDB, marked by absence of pigment (Wt area in schematic in a is shaded gray), In a: examples of non-rotated ommatidia adjacent to wt tissue are highlighted by yellow arrowheads. (b) Green arrowhead marks an under-rotated mosaic ommatidium. (c) A mutant ommatidium (yellow arrowhead) surrounded by wild-type clusters.
(d) Quantification of rotation in mosaic ommatidia: (nmoDB is shown with black bars and nmoP in white bars). (e) sev>Nmo; nmoP/nmoDB. Expression of Nmo protein in a subset of R-cells in flies homozygous mutant for nmo.