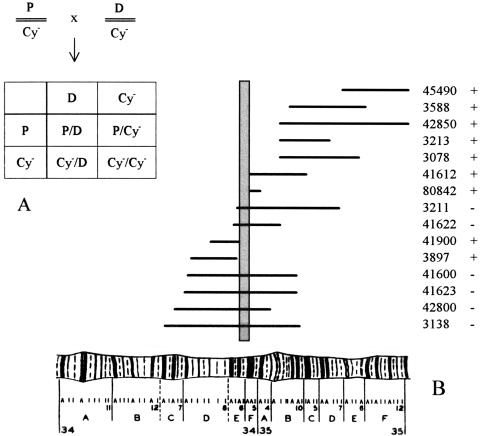
Figure 8.
Results of crossing 15 different deficiency stains (carrying a well-defined deletion in an area of chromosome 2 close to or including the DLGR-2 gene, see Table 4) with mutant P919 that carries a P element insertion in the DLGR-2 gene. (A) Schematic description of the crossings. (P) The DLGR-2 gene, carrying the P element insertion; (Cy-) curly wing mutation; (D) deletion mutation. The Cy- mutation is dominant and homozygous lethal. Therefore, if the offspring only consists of curly wing flies (P/Cy- or Cy-/D), the P/D combination is nonviable and the deletion can not be rescued by the P element-inserted DLGR-2 region. On the other hand, if the offspring contains flies with normal flat wings (P/D), the deletion mutant can be rescued. (B) Map of the deletion strains used in the rescue experiments (horizontal lines). The abscissa shows the region 34A-35F of Drosophila chromosome 2L. The stock numbers of the mutants are given at right (ordinate), together with the information about whether the deletion mutant can be rescued (+) or not (−) in a cross with mutant fly P919. For stock numbers 45490 and 42850 only the left ends of the deletions are shown, as these deletions are very large. The vertical shaded bar indicates the borders of the chormosomal region determined by the inability of mutant P919 to rescue the deletion mutants. This region is 34E5-F1, which is exactly the region where the DLGR-2 gene has been located. This is an independent and strong indication that the P element that we have earlier shown to be inserted in the DLGR-2 gene, is the only cause of lethality in mutant P919.