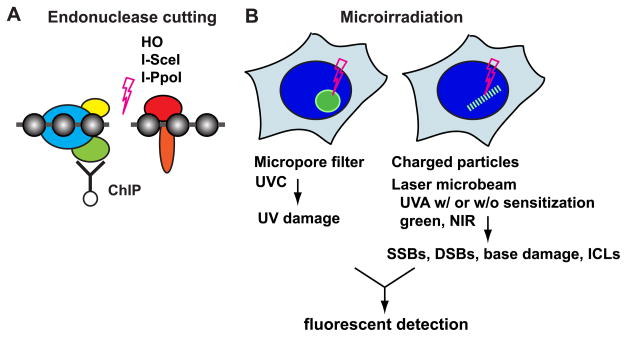
Figure 1. In vivo tools to study DNA damage recognition and response pathways.
A. ChIP analysis of endonuclease-induced DSBs. The HO endonuclease in yeast, and the I-SceI and I-PpoI homing endonucleases in mammalian cells, have been used to induce DSBs. While HO and I-PpoI cut endogenous recognition sequences, the I-SceI recognition sequence is introduced into the genome by stable transfection. Factor binding or modification in the vicinity of DSB sites can be analyzed by ChIP using specific PCR primers.
B. Fluorescent microscopy analysis following nuclear microirradiation. Selective sub-nuclear exposure of UVC using micropore filters has been used to study the NER pathway. Focused laser microbeams (of various wavelengths, energies, etc.) or charged particles, delivered to sub-micron regions in the cell nucleus (with or without DNA sensitization), have been used mainly to study DSBs. However, other types of damage are also induced, including CPD, SSBs, and in some cases, base damage. In addition, photo-activation of psoralen derivatives by microirradiation induces interstrand crosslinking damage (ICLs).