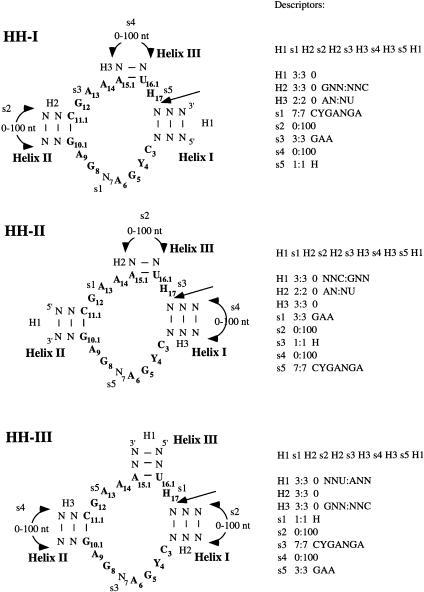
Figure 1.
Structures and descriptors of the hammerhead self-cleaving ribozyme motifs. The three descriptors, HH-I, HH-II, and HH-III, are defined by which helix is at the 5′ end and named according to the helix number (Hertel et al. 1992). Each descriptor is composed of single stranded (s) and double stranded (H) regions. The regions first are named in order from 5′ to 3′ and then specified for their length (minimum:maximum), number of mismatches (in the case of H only), and presence of specific nucleotides. For example, HH-I consists of the following features: H1 s1 H2 s2 H2 s3 H3 s4 H3 s5 H1 where H1 is an helix of a fixed length of three base pairs with no mismatches and no specific nucleotides; H2 is also of three base pairs with no mismatches but with a starting G-C base pair; H3 is an helix of 2 base pairs beginning with an A-U base pair; s1 is a single stranded region of seven nucleotides exactly with a specific sequence; s2 varies between 0 and 100 undetermined nucleotides; and so on. The hammerhead-like motifs are the same as the three shown but with an “N” replacing one of the nucleotides in boldface or with a different identity of one of the base pairs in boldface. These motifs are named according to the original motif and the position of the mutation, e.g., HH-I-3 motif is as HH-I but with an N instead of a C at position 3; thus, HH-I-3 descriptor has a modified s1 as follows: s1 7:7 NYGANGA, similarly with HH-I-iiAU, which is a HH-I motif with a A:U base pair in the Helix II instead of a G:C; thus, the descriptor HH-I-iiAU has this particularity: H2 3:3 0 ANN:NNU. The cleaving site is after H17. (H) A, C, or U; (N) A, C, G, or U; (Y) C or U. See Methods for the basis of the sequence requirements.