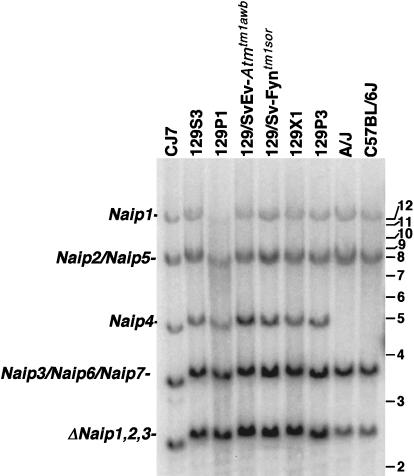
Figure 3.
Southern blot analysis of BamHI-digested genomic DNA identifies differences in Naip copy number among inbred mouse strains. Correlation of the Naip exon 11 hybridizing BamHI fragments of genomic DNA from several 129 strains with Naip loci was done by comparison of the fragments in BAC and P1 clones from the 129 haplotype that make up a complete map of the region (data not shown) and from completed genomic sequence (Endrizzi et al. 1999, 2000). Horizontal bars and numbers indicate position and size (kb) of fragments in 1-kb ladder molecular weight marker (GIBCO). A single band of 12 kb mapping to Naip1 and an 8.5-kb doublet fragment mapping to Naip2 and Naip5 are well conserved among all strains. In contrast, the 5-kb fragment mapping to Naip4 is unique to mice of the 129 lineage. A 3.8-kb band mapping to Naip3, Naip6, and Naip7 in the 129 haplotype is polymorphic in other strains. This fragment is less intense in A/J and C57BL/6J. The 2.5-kb fragment mapping to the three ΔNaip loci in the 129 haplotype is significantly decreased in intensity in A/J and C57BL/6J.