Abstract
Parafibromin, a tumor suppressor protein encoded by HRPT2/CDC73 and implicated in parathyroid cancer and the hyperparathyroidism-jaw tumor familial cancer syndrome, is part of the PAF1 transcriptional regulatory complex. Parafibromin has been implicated in apoptosis and growth arrest, but the mechanism by which its loss of function promotes neoplasia is poorly understood. We report here that a hypomorphic allele of hyrax (hyx), the Drosophila homolog of HRPT2/CDC73, rescues the loss-of-ventral-eye phenotype of lobe (Akt1s1). Such rescue is consistent with previous reports that hyx/ parafibromin is required for the nuclear transduction of Wingless/Wnt signals and that Wingless signaling antagonizes lobe function. A screen employing double hyx/lobe heterozygotes identified an additional interaction with orb and orb2, homologs of mammalian cytoplasmic polyadenylation element binding protein (CPEB), a translational regulatory protein. Hyx and orb2 heterozygotes lived longer and were more resistant to starvation than controls. In mammalian cells knockdown of parafibromin expression reduced levels of CPEB1. Chromatin immunoprecipitation demonstrated occupancy of CPEB1 by endogenous parafibromin. Bioinformatic analysis revealed a significant overlap between human transcripts potentially regulated by parafibromin and CPEB. These results show that parafibromin may exert both transcriptional and, through CPEB, translational control over a subset of target genes and that loss of parafibromin (and CPEB) function may promote tumorigenesis in part by conferring resistance to nutritional stress.
Keywords: Akt1S1, Longevity, Paf1 complex, Parafibromin, PRAS40
Introduction
Mutation of the tumor suppressor gene HRPT2/CDC73 in the germline confers susceptibility to the hyperparathyroidism-jaw tumor syndrome (HPT-JT), an autosomal dominant familial cancer syndrome with a high incidence of parathyroid malignancy (1–6). Carpten et al identified HRPT2 by positional candidate cloning (7). Somatic and/or germline inactivating HRPT2/CDC73 mutations have also been strongly implicated in sporadic parathyroid cancer (8, 9). HRPT2/CDC73 encodes parafibromin, a 531-amino acid putative tumor suppressor protein with sequence homology to Cdc73p, a yeast protein component of the RNA polymerase II-associated Paf1 complex. Recent evidence suggests that in humans parafibromin also interacts with RNA polymerase II as part of a PAF1 complex (10–12). The components of the PAF1 complex are highly conserved in Drosophila as well, including hyrax (hyx) a homolog of HRPT2 and CDC73 (13).
Despite its identification as a component of the PAF1 complex, the key molecular mechanisms by which loss of parafibromin function promotes tumorigenesis remain unclear. Endogenous parafibromin promotes apoptosis (14), and transfected parafibromin inhibits cellular proliferation (15, 16) and induces cell cycle arrest in the G1 phase (16). While these cell biological properties of parafibromin are consistent with its proposed function as a tumor suppressor, few relevant parafibromin target genes or pathways have been identified so far (17, 18). The observation, first made in Drosophila, that hyx/parafibromin binds directly to armadillo/β-catenin and facilitates Wingless (Wg)/Wnt signaling (13) has so far not given insight into the critical pathway(s) in which loss of parafibromin function leads to tumor development.
In this study we employ Drosophila as a model system to identify a genetic interaction between hyx and orb, a homolog of mammalian cytoplasmic polyadenylation element binding protein (CPEB). Flies heterozygous for hyx or orb showed enhanced longevity and marked starvation resistance. In cultured human cells RNA interference with parafibromin expression reduced the expression of CPEB1 transcript. Chromatin immunoprecipitation (ChIP) experiments demonstrated a direct association of endogenous parafibromin and other PAF1 complex components with CPEB1. Thus parafibromin may exert not only transcriptional but, through a conserved interaction with CPEB, translational control over a significant subset of its target genes.
Results
The hyrax/HRPT2 gene is essential in Drosophila development
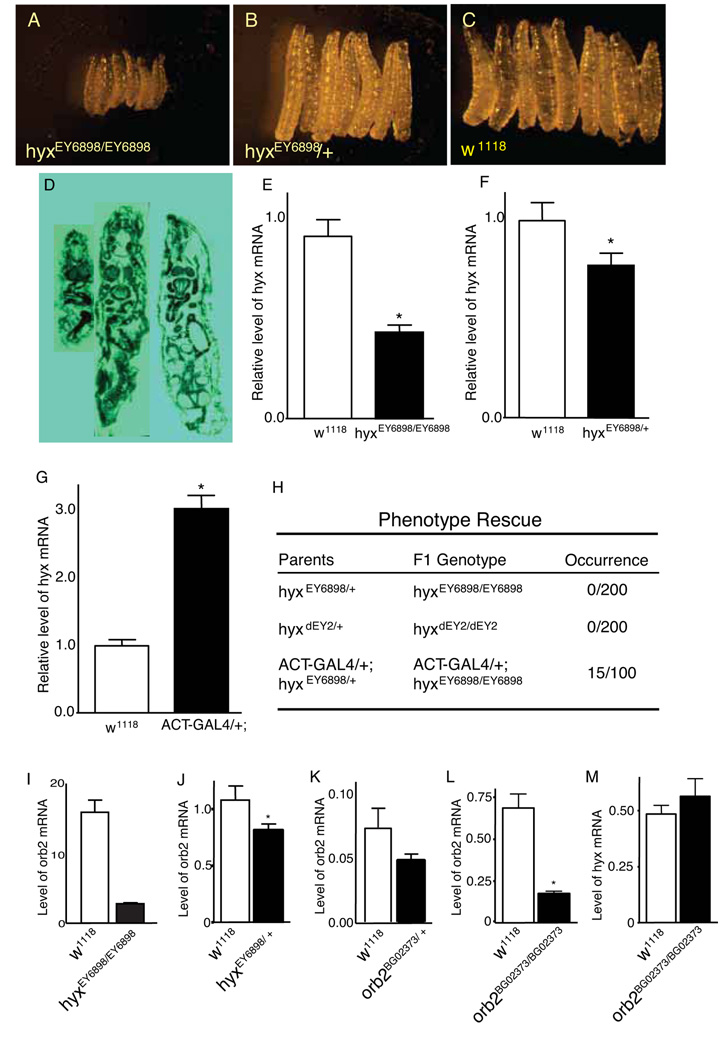
The significance of the hyx gene in Drosophila development was assessed using the hypomorphic mutant hyxEY6898. Survey of approximately 300 embryos revealed that hyxEY6898/EY6898 embryos had a developmental delay and a reduced hatching rate (22% of w1118 control embryos). Heterozygous mutant flies developed normally without obvious morphological defects but homozygotes died at early instars. Staged examination of larval development revealed that hyxEY6898/EY6898 larvae developed more slowly than both heterozygotes and w1118 control larvae. At about 48 hr after hatching, the hyxEY6898/EY6898 larvae were 1/3 the size of the dhyxEY6898/+ and w1118 controls (Fig. 1, A–C). All of the hyxEY6898/EY6898 larvae died at or before second instar. Sectioning of the 48 hr old larvae suggested that the hyxEY6898/EY6898 larvae developed disproportionately (Fig. 1D).
Fig 1. Hyrax/HRPT2 is essential for normal larval development in Drosophila and is upstream of the cytoplasmic polyadenylation element-binding protein (CPEB) homolog Orb2.
The morphology of second instar larvae were compared between hyxEY6898 homozygous and heterozygous mutants and wild type flies by stereomicroscopy (A–C) and hematoxylin and eosin histological staining (D). Shown in D., left to right, are longitudinal sections of hyxEY6898 homozygous and heterozygous mutants and w1118 larvae. The transcript levels of hyx/HRPT2 were measured by qRT-PCR in both hyxEY6898 homozygous second instar larvae (E) and heterozygous adult (F) flies. Rescue of the hyxEY6898 homozygous lethal phenotype was performed with an actin promoter-controlled GAL4 driven overexpression of the hyx/HRPT2 gene from the hyxEY6898 allele (G–H). For comparison, results with the hyxdEY2 excision mutant are also shown (H). Expression of the orb2 gene in hyxEY6898 homozygous larvae and heterozygous adult flies (I–J) and adult orb2BG02373 heterozygous and homozygous flies (K–L), and hyx/HRPT2 gene expression in orb2BG02373 homozygous mutant flies (M) was quantified by qRT-PCR. For morphology experiments, at least 50 second instar larvae were examined for each genotype. All qRT-PCR data were from at least 9 data points comprising at least three independent biological repeats.
In order to verify whether the hypomorphic hyx allele was the sole cause of the observed phenotype, we first examined the hyx gene transcript levels by quantitative RT-PCR. Compared to comparably developed hyx+/+ control flies, the hyx mRNA was approximately 55% lower in hyxEY6898/EY6898 larvae (Fig. 1E) and 26% lower in hyxEY6898/+ adult flies (Fig. 1F). Precise excision of the P element in hyxEY6898/+ flies by remobilization using a jumper line brought the hyx gene expression to the level of wild type and rescued the homozygous lethality (not shown). Furthermore the hyxEY6898/EY6898 lethal phenotype was partially rescued by overexpression of the hyx gene in the hyxEY6898/EY6898 background through genotype synthesis (19) and the UAS/GAL4 binary expression system (Fig. 1G, H.). In the presence of the 5C-actin promoter controlled GAL4 expression (act-GAL4), approximately 15% of the act-GAL4/+;hyxEY6898/EY6898 flies developed into adults while no hyxEY6898/EY6898 did (Fig. 1G, H).
Other mutant hyx alleles were tested and the penetrance of the lethality phenotype correlated inversely with the level of hyx gene expression. The hyxdEY2/+ fly was created by imprecise excision of the P-element from hyxEY6898/+. Quantitative RT-PCR analysis revealed that there was about a 40% reduction in hyx mRNA in hyxdEY2/dEY2 larvae versus w1118 controls, while three homozygous hyx deficiency mutants (hyxED5301, hyxED5331, hyxED5343) had undetectable hyx mRNA (data not shown). Although heterozygotes of all four mutants developed normally, homozygotes of the three hyx deficiency mutants died at a late embryo stage before hatching, much earlier than hyxEY6898/EY6898 flies, while the homozygotes of the imprecise excision mutant (hyxdEY2/dEY2) died in late third instar, later than the hyxEY6898/EY6898 flies. Like hyxEY6898 homozygotes, hyxdEY2/dEY2 adult flies were never found in the F1 generation of hyxdEY2/+ matings (Fig. 1H). These results confirmed the critical role of the hyx/HRPT2 gene in fly development and the GAL4-mediated partial rescue of the early larval lethality of hyxEY6898/EY6898 previously described (13).
Phenotype screening identifies orb/orb2 as hyx/HRPT2 interacting genes
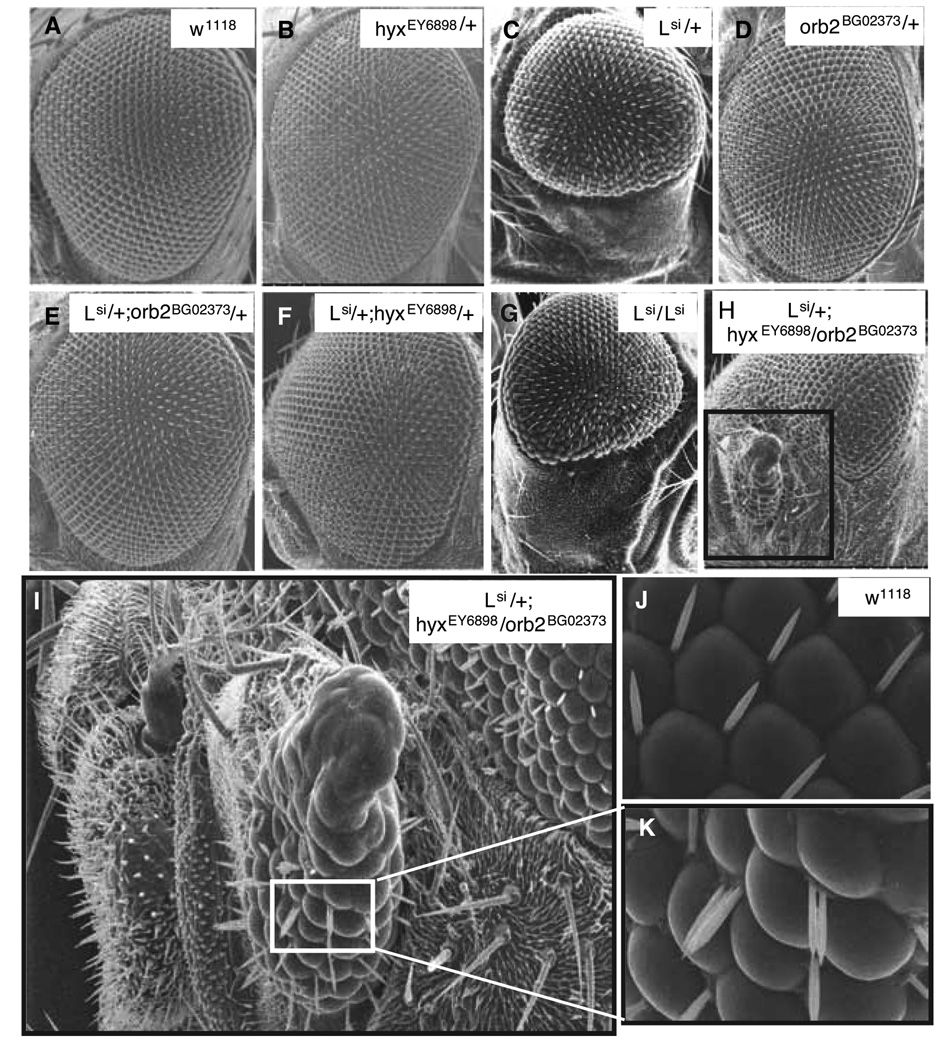
Screening for hyx suppressors or enhancers was performed by crossing hyxEY6898/+ mutants with approximately 400 selected mutant fly stocks. The fly stocks chosen for screening contained mutant alleles of fly genes homologous to human genes implicated in oncogenic, tumor suppressor or stress resistance pathways. New phenotypes distinctive from either parent were identified. It was found that doubly heterozygous mutants of hyx and lobe (L) (the homolog of mammalian Akt1 substrate1 [AKT1S1] also called the proline-rich Akt substrate of 40 kDa [PRAS40]), (Lsi/+;hyxEY6898/+), had normal eyes like hyx+/+ and hyxEY6898/+ flies (Fig. 2F, cf. 2A,B), even though heterozygous Lsi/+ flies had a loss-of-ventral-eye phenotype (Fig. 2C) (20). Thus hyx/HRPT2 could suppress or rescue the Lsi phenotype. Rescue of the half eye phenotype was confirmed by crossing hyxEY6898/+ flies with other mutant alleles of L. The half eye L mutant phenotype was rescued in all of the L and hyx double heterozygous flies generated (L1/+;hyxEY6898/+, L2/+;hyxEY6898/+, L4/+;hyxEY6898/+, L5/+;hyxEY6898/+, Lr/+;hyxEY6898/+) (not shown).
Figure 2. Genetic interaction among lobe/Akt1s1, hyx/HRPT2 and orb2/CPEB evident from Drosophila eye phenotypes.
Genetic interactions of flies were recognized by formation of novel notch and overgrowth (NOG) structures at the ventral part of eye after crosses between flies with different genotypic backgrounds. Shown are representative eye phenotypes captured by scanning electron microscopy for wild type (A), heterozygous hyxEY6898 mutant of hyx/HRPT2 (B), heterozygous Lsi mutant of lobe/AKT1S1 (C), heterozygous orb2BG02373 mutant of orb2/CPEB (D); double heterozygous mutants of lobe and orb2 (E), lobe and hyx/HRPT2 (F); homozygous lobe Lsi mutant (G) and triple heterozygous hybrid mutants of lobe, hyx/HRPT2 and orb2 genes (H). Higher magnification image of boxed region of H is shown in I. The ommatidia and sensory bristle phenotype of the triple heterozygous lobe, hyx/HRPT2 and orb2 mutant are shown (I, K). Higher magnification image of boxed region of I is shown in K, with wild type shown for comparison (J).
The basis of the genetic interaction observed between hyx/HRPT2 and lobe is unknown. Since Lsi is a likely dominant negative mutation with high penetrance (21) and hyxEY6898/+ heterozygotes had no obvious morphological phenotype, we hypothesized that the combination of the Lsi/+ and hyxEY6898/+ mutations might sensitize the genetic background to allow rapid identification of genes that interact with L or hyx/HRPT2 or both. To explore this possibility we employed a doubly heterozygous mutant fly line (Lsi/cyo; hyxEY6898/Tm3) generated by genotype synthesis (19). We first tested the hypothesis by crossing the lobe/hyx double heterozygotes with strains carrying mutations of PAF1 complex component genes besides hyx, expected to interfere with the rescue of L by hyx.
To this end the doubly heterozygous mutant fly ( Lsi/+; hyxEY6898/+ ) was crossed with fly strains harboring mutations in PAF1 complex component homologs, including atms/paf1(CG2503), atu/leo1(CG1433) and dctr9 (CG9899) genes (Table I, crosses 11–14). The triply heterozygous flies Lsi/+;hyxEY6898/atus1938/+ and Lsi/+;hyxEY6898/dctr9NP5197/+ had a high percentage of flies with a distinctive, novel eye phenotype ranging from notched ventral eye field with small overgrowths to a half eye with large dysplastic overgrowths present in the missing ventral eye region (NOG phenotype) (Table I, crosses 11,14). In contrast, doubly heterozygous offspring from the same crosses with genotypes Lsi/+;atu10217/+ and Lsi/dctr9NP5197 had normal eyes (Table I, crosses 11,14). Two mutant alleles of the Paf1 homolog atms were employed, atmsNP5451 and atmsrk509. Both the Lsi/+;hyxEY6898/atmsNP5451 and the Lsi/+;hyxEY6898/atmsrk509 triply heterozygous offspring expressed the NOG phenotype but the latter had a higher penetrance (Table I, crosses 12,13). However some ~20% of the L324/+;atmsrk509/+ double heterozygotes also had NOG eyes, indicating there might be a hyx-independent interaction between atms/Paf1 and Lsi genes (Table I, cross 13). Similarly, genetic interaction evidenced by the NOG phenotype was also observed between Lsi/+;hyxEY6898/+ and the Armadillo(Arm)/β-catenin genes (Table I, cross 20) (13).
Table I.
Genetic Interactions of Selected Genes
| Parents | F1 Genotype | % Flies with NOGa eye phenotype |
Molecular Function |
||
|---|---|---|---|---|---|
| Parent 1 | Parent 2 | No. of Crosses |
|||
| Lsi/+ | W1118 | 1 | Lsi/+b | 0 | L is the fly homolog of mammalian AKT1S1 |
| hyxEY6898/+ | 2 | Lsi/+; hyxEY6898/+ | 0 | Hyx is the fly homolog of parafibromin, a component of PAF1 complex associated with RNA polymerase II |
|
| hyxEY6898/+ | W1118 | 3 | hyxEY6898/+ | 0 | |
| Lsi/+;hyxEY6898/+ | orb2BG02373/+ | 4 | Lsi/+;hyxEY6898/orb2BG02373 | 23 | Orb and Orb2 are homologs of human CPEB, a component of the eIF-4E translation initiation complex, regulating polyadenylation of certain mRNAs |
| Lsi/+;orb2BG02373/+ | 6 | ||||
| orb2Scf-R6/+ | 5 | Lsi/+;hyxEY6898/orb2Scf-R6 | 26 | ||
| Lsi/+;orb2Scf-R6/+ | 4 | ||||
| orb2Scf-R11/+ | 6 | Lsi/+;hyxEY6898/orb2Scf-R11 | 12 | ||
| Lsi/+;orb2Scf-R11/+ | 0 | ||||
| orbdec/+ | 7 | Lsi/+;hyxEY6898/orbdec | 2 | ||
| Lsi/+;orbdec/+ | 0 | ||||
| orbEY08547/+ | 8 | Lsi/+;hyxEY6898/orbEY08547 | 0 | ||
| Lsi/+;orbEY08547/+ | 0 | ||||
| orbExel6274/+ | 9 | Lsi/+;hyxEY6898/orbExel6274 | 21 | ||
| Lsi/+;orbExel6274/+ | 0 | ||||
| orbExel6280/+ | 10 | Lsi/+;hyxEY6898/orbExel6280 | 6 | ||
| Lsi/+;orbExel6280/+ | 0 | ||||
| atus1938/+ | 11 | Lsi/+;hyxEY6898/atus1938 | 33 | Atu, Atms, and dCtr9 are the fly homologs of Leo1, Paf1 and Ctr9 respectively, components of the PAF1 complex |
|
| Lsi/+;atus1938/+ | 0 | ||||
| atmsNP5451/+ | 12 | Lsi/+;hyxEY6898/atmsNP5451 | 23 | ||
| Lsi/+;atmsNP5451/+ | 0 | ||||
| atmsrk509/+ | 13 | Lsi/+;hyxEY6898/atmsrk509 | 46 | ||
| Lsi/+;atmsrk509/+ | 19 | ||||
| dctr9NP5197/+ | 14 | Lsi/+;hyxEY6898/dctr9NP5197 | 25 | ||
| Lsi/+;dctr9NP5197/+ | 0 | ||||
| ArmG0192 | 20 | ArmG0192/+;Lsi/+;hyxEY6898/+ | 12 | Arm (Armadillo) is WNT signaling pathway component |
|
| ArmG0192/+;Lsi/+;+/+ | 0 | ||||
| 4EBPk10101 | 21 | Lsi/4EBPk10101;hyxEY6898/+ | 0 | Components of Foxo signaling pathway |
|
| Lsi/4EBPk10101;+/+ | 0 | ||||
| 4EBP06270 | 22 | Lsi/4EBP06270;hyxEY6898/+ | 0 | ||
| Lsi/4EBP06270;+/+ | 0 | ||||
| 4EBPk13506 | 23 | Lsi/4EBPk13506;hyxEY6898/+ | 0 | ||
| Lsi/4EBPk13506;+/+ | 0 | ||||
| 4EBPEY00763 | 24 | Lsi/4EBPEY00763;hyxEY6898/+ | 0 | ||
| Lsi/4EBPEY00763;+/+ | 0 | ||||
| FoxoED5634 | 25 | Lsi/+;hyxEY6898/ FoxoED5634 | 0 | ||
| Lsi/+; FoxoED5634/+ | 0 | ||||
| FoxoDG02610 | 26 | Lsi/+;hyxEY6898/FoxoDG02610 | 0 | ||
| Lsi/+;Foxo DG02610/+ | 0 | ||||
| FoxoEY16506 | 27 | Lsi/+;hyxEY6898/FoxoEY16506 | 0 | ||
| Lsi/+;FoxoEY16506 /+ | 0 | ||||
| FoxoBG01018 | 28 | Lsi/+;hyxEY6898/FoxoBG01018 | 0 | ||
| Lsi/+;Foxo BG01018/+ | 0 | ||||
| FoxoEY11248 | 29 | Lsi/+;hyxEY6898/FoxoEY11248 | 0 | ||
| Lsi/+;Foxo EY11248/+ | 0 | ||||
| FoxoED5644 | 30 | Lsi/+;hyxEY6898/FoxoED5644 | 0 | ||
| Lsi/+; FoxoED5644/+ | 0 | ||||
NOG: Notch and Overgrowth, with notches and variably sized dysplastic cornlike overgrowths in the ventral eye fields
Typical Lsi/+ phenotype is absence of ventral eye fields
Using the same strategy we crossed the doubly heterozygous (Lsi/+; hyxEY6898/+) flies with other individual heterozygous flies from the collection of stock mutants strains to screen for new genetically interacting genes. The genetically interacting genes were defined as those that produced novel eye phenotypes in one of three possible triple heterozygous genotypes: +/+; Lsi/X; hyxEY6898/+ or +/+; Lsi/+; hyxEY6898/X or X/+; Lsi/+; hyxEY6898/+, where X was the tested gene mutant. Approximately 5% of the ~400 screened gene mutant strains produced abnormal eye phenotypes (see below, and manuscript in preparation).
Crosses of Lsi/+; hyxEY6898/+ double heterozygotes with flies harboring a mutant allele of the cytoplasmic polyadenylation element binding protein (CPEB) homolog orb2, orb2BG02373, produced offspring with a high frequency of the NOG phenotype (Fig. 2H, I, K). CPEB is an RNA-binding zinc-finger protein that controls the cytoplasmic polyadenylation of certain mRNAs and can repress or mask the their translation (22). Approximately 25% of the triple heterozygotes (Lsi/+;hyxEY6898/orb2BG02373) flies had NOG eye phenotypes including half eye with large overgrowths in the missing ventral eye region (Fig. 2H, I, K; Table I, cross 4). Although the dysplastic tissue frequently had a recognizable eye-like arrangement, the ommatidia and sensory bristles were deformed and in disarray (Fig. 2K, cf. 2J). Testing of two other orb2 mutant alleles gave similar results (Table I, crosses 5, 6). Although crossing the Lsi/+; hyxEY6898/+ double heterozygotes with the orb gene mutants did produce a similar NOG phenotype, the penetrance was lower than that seen in the orb2 crosses (Table I, crosses 7–10). In contrast ~95% of the doubly heterozygous mutant flies (Lsi/+;orb2BG02373/+, Lsi/+;orbdec, Lsi/+;hyxEY6898/+, hyxEY6898/orb2BG02373/+ and hyxEY6898/orbdec) had normal eyes (Fig. 2E, F; Table I, crosses 4–7). These interactions were further confirmed by similar interactions of Lsi/+;hyxEY6898/+ with the deficiency mutants of both orb2 and orb (Table I, crosses 5,6,9,10). Because all other possible doubly heterozygous mutants from the same matings produced flies with normal eyes (Table I, crosses 5,6,9,10), the observed NOG eye phenotype must be from the specific combination of hyx and orb loss of function. As noted above, the triply heterozygous Lsi/+;hyxEY6898/orb2 mutant flies produced significantly larger overgrowths and exhibited higher penetrance of the NOG phenotype than the Lsi/+;hyxEY6898/orb flies, suggesting a stronger interaction of hyx with the orb2 alleles.
Because the doubly heterozygous mutant flies (Lsi/+;hyxEY6898/+) only genetically interacted with ~5% of our collection of mutant fly stocks, it is reasonable to believe that the genetic interactions with orb and orb2 are specific. To further assess the selectivity of the interactions, we also tested to see if the Lsi/+;hyxEY6898/+ double heterozygotes interacted with the foxo and 4EBP genes (Table I, crosses 21–30). Like L/akt1s1, Foxo is a downstream target of the akt gene. The protein 4EBP competes with eIF-4G for binding to eIF-4E to regulate translational initiation (23), a process that CPEB can also regulate through interaction with the eIF-4E-binding protein Maskin (24). The Lsi/+;hyxEY6898/+ flies were crossed with six mutants of foxo and four mutants of thor (4EBP homolog). No genetic interactions were observed among the resulting triply heterozygous flies (Table I, crosses 21–30).
The eye phenotype in Lsi/+;hyxEY6898/orb2BG02373 triple heterozygotes is associated with an abnormal pattern of apoptosis and increased ectopic cell proliferation in larval eye discs
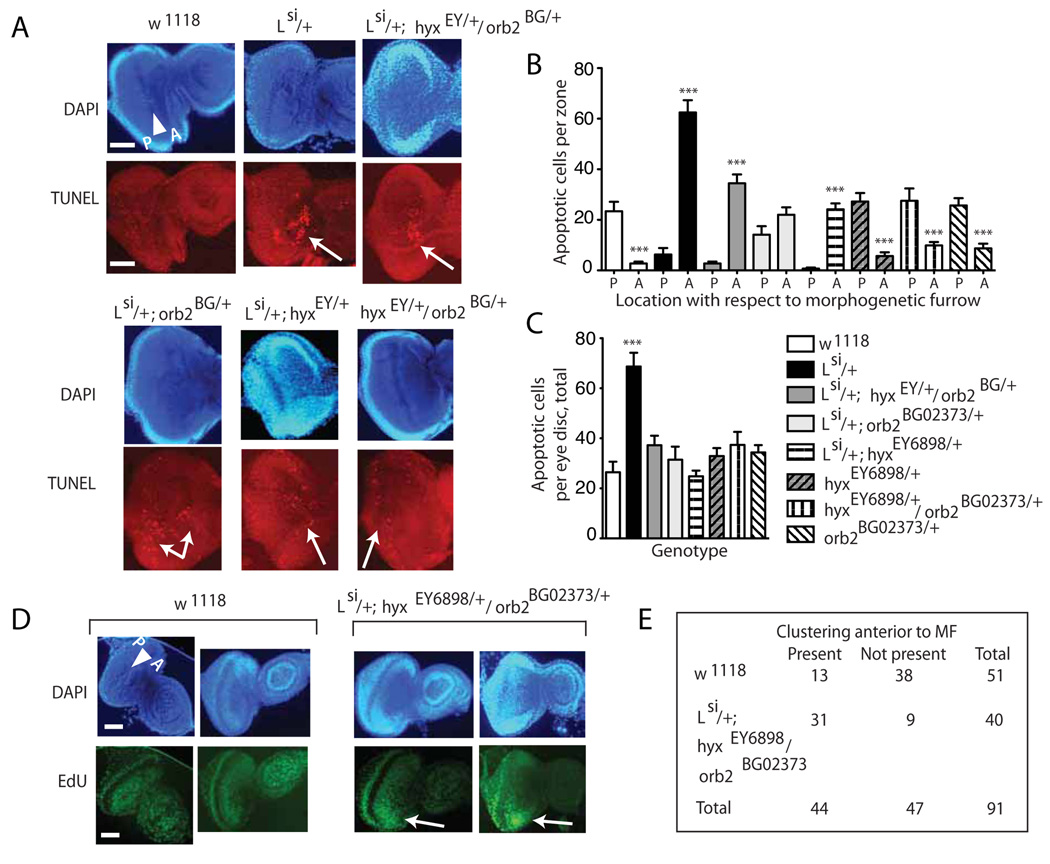
The NOG phenotype observed in the Lsi/+;hyxEY6898/+/orb2BG02373/+ triple heterozygotes could result from either impaired apoptosis and/or excessive proliferation during eye development. The imaginal discs of third instar larvae of wild-type and Lsi/+;hyxEY6898/+/orb2BG02373/+ triple heterozygote flies were therefore examined using the TUNEL assay to identify the nuclei of cells undergoing apoptosis, and compared to discs of Lsi/+ larvae, since loss of L has been shown to induce apoptosis at this stage (20), as well as to discs from larvae from the three double heterozygote combinations and the other single heterozygotes (Fig. 3A–C). In the developing imaginal disc, the morphogenetic furrow (MF) moves across the eye field in the posterior to anterior direction, inducing differentiation in the cells directly posterior to it (25). The majority of TUNEL-positive apoptotic cell nuclei in the eye imaginal discs of wild-type, hyxEY6898/+, orb2BG02373/+, and double heterozygote hyxEY6898/+;orb2BG02373/+ eye discs are located posterior to the MF and uniformly distributed whereas the majority of apoptotic nuclei in the Lsi/+ single heterozygotes and Lsi/+; hyxEY6898/+/orb2BG02373/+ triple heterozygotes are found anterior to the MF and grouped in large clusters (Fig. 3A, B). The majority of apoptotic nuclei in the Lsi/+; hyxEY6898/+ double heterozygotes are also localized anterior to the MF, but are dispersed rather than grouped. There was no significant difference with respect to the MF in the location of apoptotic nuclei in Lsi/+; orb2BG02373/+ double heterozygotes (Fig. 3A, B). The total number of TUNEL-positive apoptotic nuclei per entire eye imaginal disc of Lsi/+ heterozygote larvae was significantly greater than either w1118 control, triple, double, or other single heterozygote larvae (Fig. 3C). Cell proliferation in the larval eye discs was assayed by the incorporation of the synthetic nucleoside 5-ethynyl-2´-deoxyuridine (EdU) into newly synthesized DNA. In wild-type larvae, EdU incorporation was evenly distributed anterior to the MF, where cells are cycling asynchronously, and was slightly more intense just posterior to the MF, where cells are undergoing synchronous S phase (Fig. 3D). In the Lsi/+; hyxEY6898/orb2BG02373 triple heterozygotes, staining for EdU incorporation frequently revealed bright clusters of proliferating cells anterior to the MF suggesting an increase in ectopic cellular proliferation. Thus the developing eye discs of Lsi/+; hyxEY6898/orb2BG02373 triple heterozygotes with a propensity to later develop the NOG phenotype were characterized by both an abnormal pattern of apoptosis and an increased incidence of ectopic cellular proliferation.
Figure 3. The imaginal eye disc of triply heterozygous mutant Lsi/+; hyxEY6898/orb2BG02373 larvae is characterized by an abnormal pattern of apoptosis and increased ectopic cellular proliferation.
A. DAPI nuclear staining (upper) and TUNEL analysis (lower) of the eye imaginal discs of wild-type, Lsi/+, Lsi/+; hyxEY6898/+/orb2BG02373/+ triple heterozygote, and Lsi/+; hyxEY6898/+Lsi/+; orb2BG02373/+, and hyxEY6898/+;orb2BG02373/+ double heterozygote third instar larvae. White triangle indicates the morphogenetic furrow (MF), with anterior (A) and posterior (P) directionality indicated. Scale bar = 1 mm. Please note that in the legend to A. (and in the legend in C. for the triple heterozygote) the genotype for hyxEY6898/+ is abbreviated hyxEY/+ and the genotype for orb2BG02373/+ is abbreviated to orb2BG/+B. The majority of apoptotic cell nuclei in the wild-type, hyxEY6898/+orb2BG02373/+, and hyxEY6898/+/orb2BG02373/+ eye discs were located posterior (P) to the MF and uniformly distributed whereas the majority of apoptotic nuclei in the Lsi/+ heterozygotes and Lsi/+; hyxEY6898/+/orb2BG02373/+ triple heterozygotes are found anterior (A) to the MF and grouped in large clusters (white arrows in figure part A.). The majority of apoptotic nuclei in the Lsi/+; hyxEY6898/+ double heterozygotes are also localized anterior to the MF, but are dispersed rather than grouped (***, p <0.0001, anterior vs. posterior, 2-tailed t test). There was no significant difference in the anterior and posterior distribution of apoptotic nuclei in Lsi/+; orb2BG02373/+ double heterozygotes (p=0.08, anterior vs. posterior, 2-tailed t test). Legend as in C. C. The total number of TUNEL-positive apoptotic nuclei per eye disc in Lsi/+ heterozygotes is significantly increased compared to wild-type, triple, double and other single heterozygotes (***, p <0.0001, Lsi/+ vs. wt or double, triple or other single heterozygotes, 2-tailed t test). For B. and C. the number of distinct eye imaginal discs counted: wt, n= 20; Lsi/+, n= 22; Lsi/+; hyxEY6898/+/orb2BG02373/+ triple heterozygotes, n= 42; Lsi/+; orb2BG02373/+, n=19; Lsi/+; hyxEY6898/+, n=20; hyxEY6898/+, n=16; orb2BG02373/+, n=16; hyxEY6898/+; orb2BG02373/+, n=16. D. Third instar larvae eye discs of wild-type and Lsi/+; hyxEY6898/orb2BG02373 triple heterozygotes with nuclei stained with DAPI (upper) and proliferating cells stained for the incorporation of the nucleoside 5-ethynyl-2´-deoxyuridine (EdU) (lower) as described in Materials and Methods. Labels in DAPI images as in A. White arrows indicate bright clusters of proliferating cells anterior to MF. E. 2 × 2 contingency table showing the number of wild-type and triple heterozygote eye discs in which bright clusters of proliferating cells anterior to the MF were observed, scored as described in Materials and Methods (n = 51 for wild-type, n = 40 for triple heterozygotes; Fisher’s exact test, 2-tailed p value <0.0001).
The hyx/HRPT2 and orb2 genes regulate fly lifespan and starvation resistance
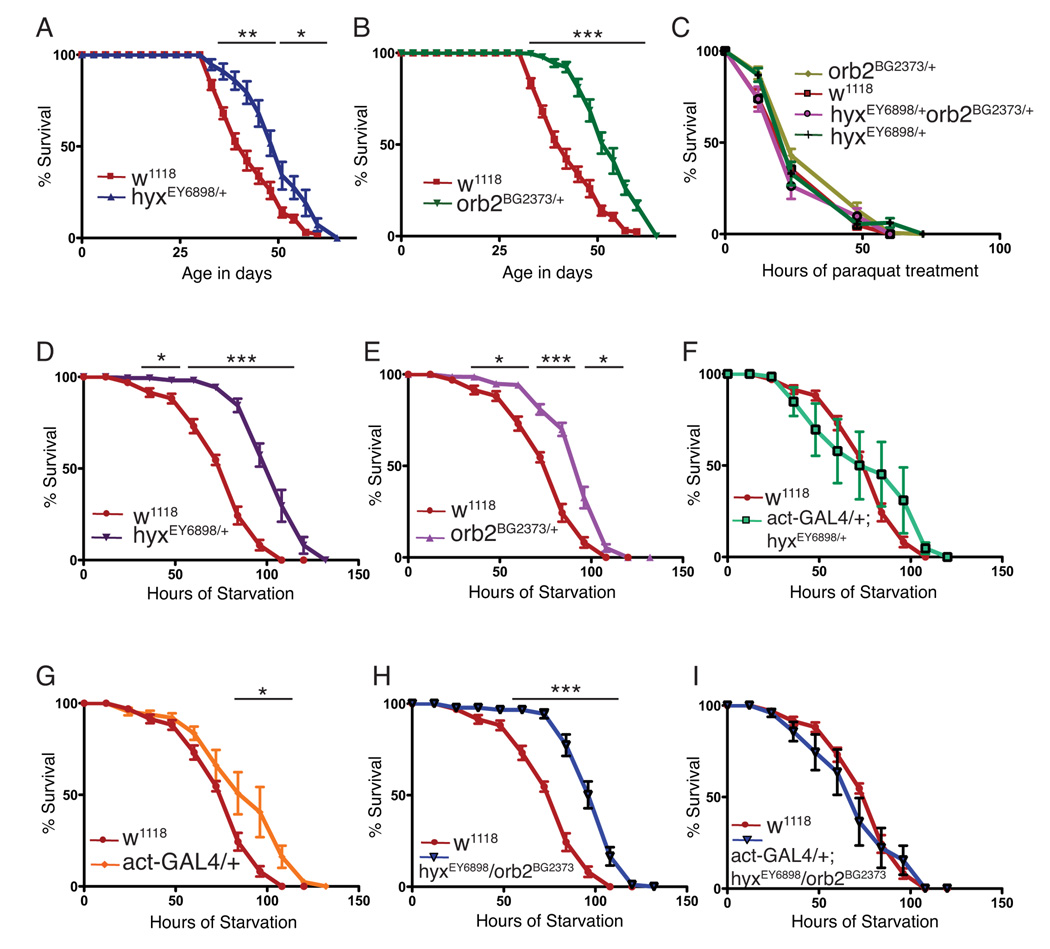
HRPT2/CDC73 is a putative tumor suppressor gene (7). Well characterized tumor suppressor genes like P53 and Rb have been implicated in the processes of cellular senescence, longevity and stress resistance (26). In addition CPEB has been shown to regulate cellular senescence in mouse embryonic fibroblasts (22). We therefore tested if hyx/HRPT2 and/or orb2 might control longevity and stress resistance in the present fly model. Flies heterozygous for hyx/HRPT2 or orb2 had significantly enhanced longevity compared to w1118 control flies (Fig. 4A, B). Since longer lifespan is often associated with enhanced stress resistance in fly, we tested if oxidative stress resistance was a factor causing longer lifespan in both hyx/HRPT2 and orb2 heterozygous mutants. However no significant differences in resistance to paraquat (an in vivo free radical generator) treatment were observed, suggesting that oxidative stress is not the cause of the extended lifespan (Fig. 4C).
Figure 4. Enhanced longevity and starvation resistance in hyx/HRPT2 and Orb2/CPEB mutant flies.
The longevity of the indicated heterozygous hyx /HRPT2 and orb2 mutant flies were examined under standard culture conditions as compared to wild type flies (A and B). Survival upon exposure to the herbicide and oxygen free radical–generator paraquat of wild-type and the indicated mutant flies is shown (C). Flies in C were fed with a paraquat-sucrose solution. Survival upon starvation (D–I) of the indicated single or double hyx/HRPT2 and orb2 heterozygous mutants is shown. Flies in D-I were supplied only with water to test starvation resistance. Experiments aimed at the rescue of the hyxEY6898 enhancer trap mutant by mating with a driver strain expressing GAL4 from the 5C-actin promoter (act-GAL4) are shown in F and I, with the driver-only control shown in G. The number of surviving flies was recorded every three days for lifespan tests and daily for stress tests. Ten or more vials were used for each experiment and 3 or more independent experiments were conducted for each fly line. Vials contained 20 flies each for lifespan determination and 10 flies each for stress tests. Each data point shown represents the pooled mean survival from 10 to 12 vials of the indicated genotype, except for the w1118 flies used in stress testing in which each data point represents the pooling of 20 vials (*, p <0.05; **, p <0.001; ***, p <0.0001; vs. wt for the indicated time points, 2-tailed t test).
Resistance to starvation however was found to be significantly higher in both hyx/HRPT2 (hyxEY6898/+) and orb2 (orb2BG2373/+) heterozygous flies compared to controls (Fig. 4D–E). To prove that the hyx/HRPT2 mutation was the cause of the extended lifespan phenotype, we tested for rescue of the phenotype upon overexpression of the hyx/HRPT2 gene on the hyxEY6898 allele in act-GAL4/+;hyxEY6898/+ flies. As shown in Fig. 4F, overexpression of hyx/HRPT2 gene restored starvation sensitivity to near control levels, whereas flies with 5C-actin promoter-driven GAL4 expression only (but lacking the GAL4-sensitive hyxEY6898 allele) had slightly enhanced starvation resistance (Fig. 4G). Note that the incomplete rescue of the lethality phenotype by actin GAL4-driven hyx/HRPT2 expression documented in Fig. 1H was against a hyxEY6898/ EY6898 background whereas the full rescue of the starvation resistance phenotype by actin GAL4-driven hyx/HRPT2 overexpression in Fig. 4F was in the context of hyxEY6898/+ flies. Furthermore, flies doubly heterozygous for hyx/HRPT2 and orb2 (hyxEY6898/+/ orb2BG2373/+) showed starvation resistance similar to the hyxEY6898/+ flies (Fig. 4H, cf. 4D). Doubly heterozygous hyxEY6898/+/ orb2BG2373/+ flies with 5C-actin promoter-driven GAL4 overexpression of hyx/HRPT2 from the hyxEY6898 allele exhibited starvation resistance comparable to control flies (Fig. 4I, cf. 4H), suggesting that hyx/HRPT2 gene overexpression could also blunt the effects of orb2.
Hyrax/HRPT2 functions upstream of orb2
The genetic interaction between hyx/HRPT2 and orb2 observed in the eye phenotypes of the L triple heterozygotes, as well as the lack of synergy in the lifespan and starvation resistance experiments, suggest that hyx/HRPT2 and orb2 function in the same pathway, but provide no insight into the relative position of the two genes. To understand their functional relationship better, quantitative RT-PCR was used to measure transcript levels of the genes in fly strains carrying mutant alleles for hyx/HRPT2 or orb2. Levels of orb2 transcript were reduced by some 80% in larvae of hyxEY6898 homozygotes and by 25% in adult hyxEY6898 heterozygotes compared to control (Fig. 1I, J). In contrast, adult flies homozygous for orb2BG02373 had normal levels of hyx/HRPT2 transcript (Fig. 1M) even though orb2 message levels were successively reduced in heterozygous and homozygous orb2BG02373 flies (Fig. 1K, L). Taken together these results indicate hyx/HRPT2 functions upstream of orb2 acting at least in part at the transcriptional level.
Parafibromin regulates the transcription of CPEB isoforms in mammalian cells
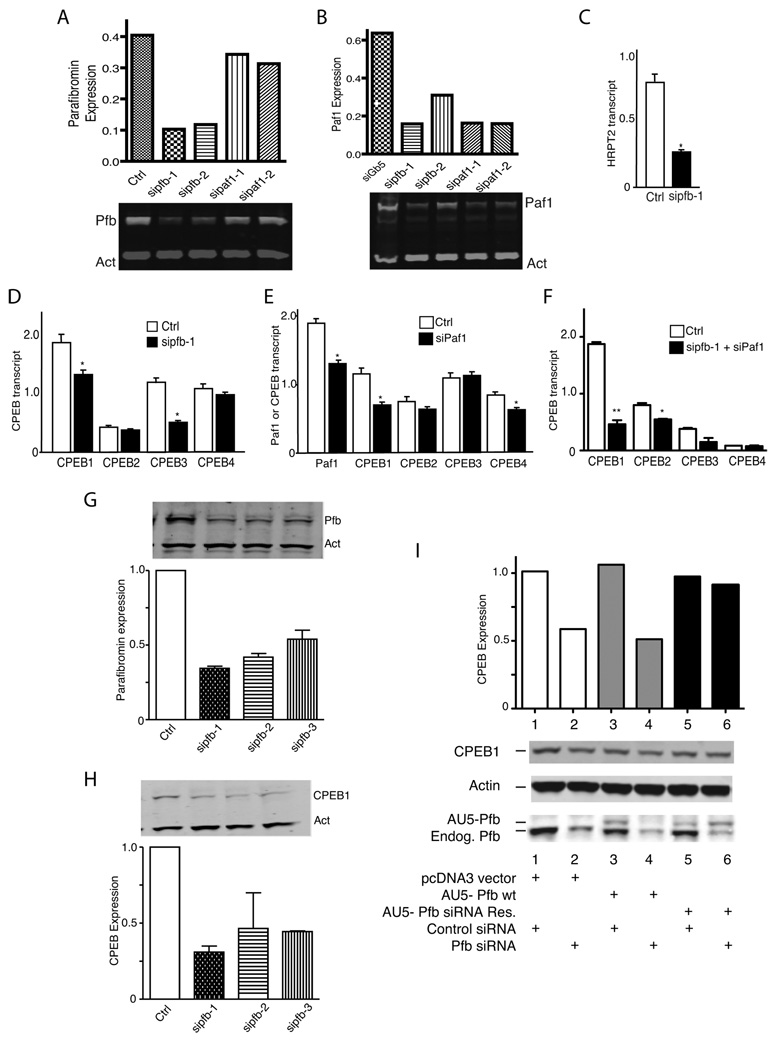
Hyx and parafibromin represent a family of gene products widely conserved among eukaryotes as components of Paf1/PAF1 complexes. We therefore examined mammalian cells for evidence of a conserved pathway connecting HRPT2/CDC73 and CPEB comparable to that between hyx/HRPT2 and orb2. To study this we employed RNA interference to impair the expression of parafibromin and Paf1 (14, 17). Transfection of HEK-293FT cells with small interfering duplex RNAs (siRNA) targeting two different sequences of the HRPT2/CDC73 or Paf1 transcripts inhibited the expression of their target genes compared to scrambled siRNA (Ctrl) or siRNA targeting the unrelated gene Gβ5, as evidenced by quantitative immunoblotting (Fig. 5A,B). Knockdown of parafibromin expression also impaired the expression of Paf1 in these cells (Fig. 5B). RNA interference employing HRPT2- and Paf1-targeted siRNAs was therefore used to study the effect of parafibromin and Paf1 knockdown on mammalian CPEB expression.
Figure 5. Knockdown of parafibromin impairs CPEB1 expression at the transcriptional level.
The expression of HRPT2, Paf1 and CPEB1-4 genes in human embryonic kidney cells after RNA interference employing siRNAs targeting HRPT2 (sipfb) and Paf1 (sipaf1) as analyzed by immunoblotting using infrared imaging or quantitative RT-PCR is shown. (A) and (B): expression of parafibromin and Paf1 protein by immunoblot (lower panels) and quantification the indicated bands normalized to the actin (Act) loading control by infrared imaging (upper). (C), (D), (E) and (F): transcript levels of the HRPT2, Paf1, and CPEB1-4 genes in HRPT2- and/or Paf1-siRNA treated and control siRNA-treated cells were measured by quantitative RT-PCR. (*, p <0.05; **, p <0.005 vs. control transcript level, 2-tailed t-test) (G) and (H): immunoblot analysis of parafibromin (Pfb) and CPEB1 protein expression in control or HRPT2-siRNA treated cells (insets) with lower histograms showing quantification of expression relative to actin based on infrared imaging of immunoblots. (I): expression of CPEB1 in cells transfected with the empty pcDNA3 vector only, wild-type AU5 epitope-tagged parafibromin cDNA, AU5 epitope-tagged parafibromin cDNA engineered with silent base changes to render it resistant to siPfb-1 siRNA, and either control siRNA or siPfb-1 siRNA, as indicated, was determined by immunoblot (lower panels) and quantified relative to actin, by infrared imaging of immunoblots (upper graph). Experiments shown in A, B, G and H used HEK-293FT cells, while the experiments shown in C, D, E, F, and I used HEK-293 cells. For qRT-PCR each repeat employed triplicate reactions and each data set represents an n= 9 or more. All experiments are representative of three or more independent biological repeats.
There are four CPEB isoforms in mammals, CPEB1-4. RNA interference targeting the HRPT2/CDC73 transcript knocked down HRPT2/CDC73 transcript levels, as expected (Fig. 5C), and significantly diminished the expression of CPEB1 and CPEB3 but had little effect on CPEB2 and CPEB4 (Fig. 5D). If the effects of HRPT2/parafibromin knockdown on CPEB involved the former’s role as a component of the PAF1 complex, then knocking down Paf1 might also affect CPEB gene expression. Reduction of Paf1 gene expression by RNA interference significantly reduced CPEB1 and CPEB4 expression but had no effect on CPEB2 or CPEB3 (Fig. 5E). To check if the effects of parafibromin and Paf1 knockdown on CPEB expression were redundant or additive, we treated cells with both parafibromin and Paf1 siRNAs and measured the resulting transcript levels of the CPEB isoforms (Fig. 5F). Combined interference with both parafibromin and Paf1 expression enhanced the knockdown of CPEB1 and CPEB2 expression but had little additional effect on CPEB3 or CPEB4 (Fig. 5F). The additivity of effect on CPEB1 and CPEB2 transcript levels could argue against the involvement of the PAF1 complex or, since treatment with parafibromin and Paf1 siRNAs only partially knocked down expression of their cognate proteins (see. Fig. 5A, B), may alternatively reflect a synergistic reduction of PAF1 complex expression and function when both parafibromin and Paf1 siRNAs were employed. Taken together, these results suggested that while parafibromin and Paf1 co-regulate CPEB1 gene expression through a mechanism that may involve the PAF1 complex, regulation of CPEB3 and CPEB4 gene expression might be through alternative pathways.
Quantitative immunoblots of parafibromin and CPEB1 were performed after RNA interference with parafibromin expression using siRNAs targeting three different regions of the HRPT2/CDC73 transcript (Fig. 5G, H). All three HRPT2/CDC73-directed siRNAs reduced parafibromin expression and also knocked down CPEB1 protein levels, with the most effective reagent being the sipfb-1 siRNA (Fig. 5G, H). To test the specificity of the RNA interference, silent base changes designed to impair interaction with sipfb-1 siRNA were introduced into a cDNA encoding an AU5-epitope-tagged human parafibromin and a rescue experiment performed (Fig. 5I). Whereas CPEB1 expression was knocked down by sipfb-1 siRNA treatment it was rescued by transfection of siRNA-resistant AU5-parafibromin, but not wild-type AU5-parafibromin (Fig. 5I). Rescue of CPEB1 expression was seen with two other siRNA-resistant AU5-parafibromin mutants with different silent base changes (not shown). The sipfb-1 siRNA treatment was effective at impairing the expression of both endogenous parafibromin and transfected wild-type AU5-parafibromin (with slightly slower mobility on SDS-PAGE due to the epitope tag), but consistently enhanced expression of the siRNA-resistant AU5-parafibromin (Fig. 5I, parafibromin immunoblot panel, cf. lanes 5,6 vs. lanes 1,2 and 3,4). We speculate that this paradoxical effect may be due to increased stability of the AU5-parafibromin protein encoded by the siRNA-resistant cDNA when it is able to complex with other PAF1 complex components, an ability that is enhanced when competing endogenous parafibromin is knocked down.
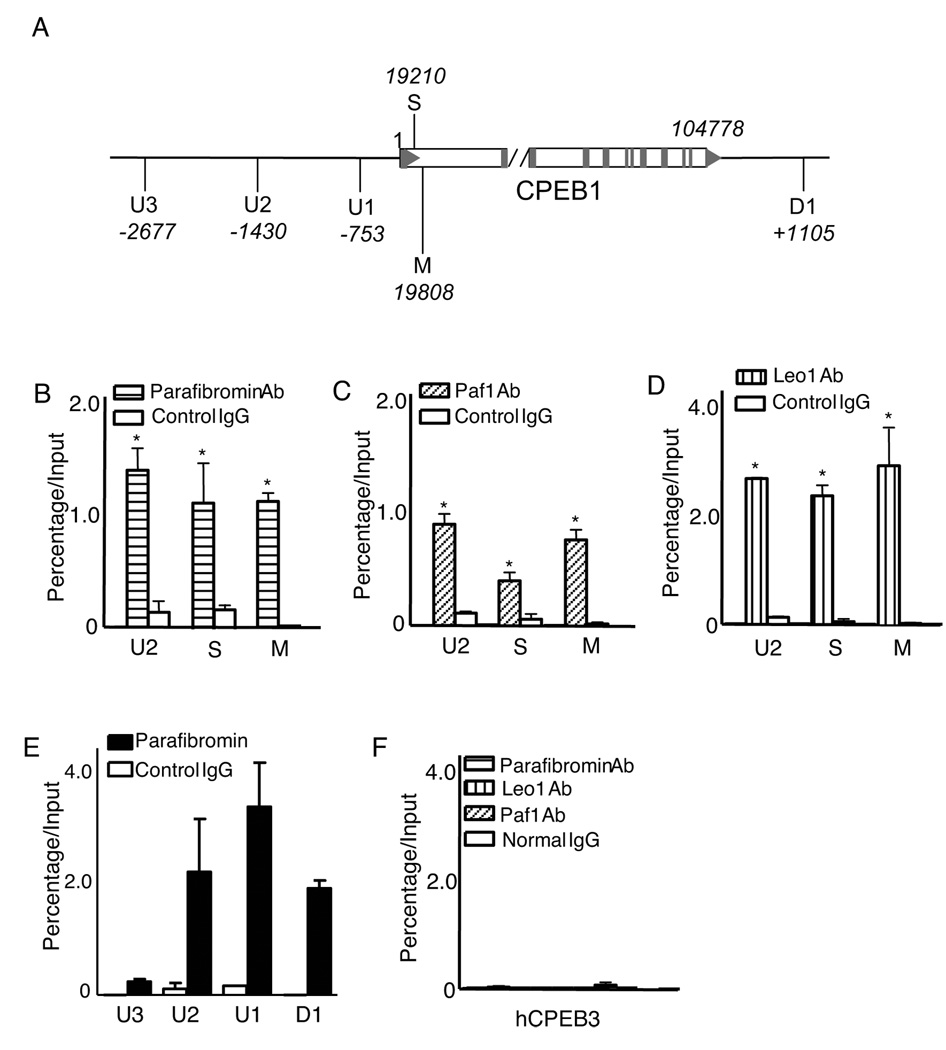
Parafibromin and other PAF1 complex components bind to the CPEB1 gene
Knockdown of parafibromin reduced CPEB1 mRNA and protein levels. ChIP was used to determine if this effect was consistent with regulation at the transcriptional level. Six pairs of primers were used to interrogate anti-parafibromin immunoprecipitates in the ChIP assay by quantitative PCR, three sets targeting regions upstream of the CPEB1 transcriptional start site (U1, U2, and U3), two sets targeting internal gene coding sequences (S and M), and one set directed at a region 1100 bp downstream of the CPEB1 gene (D1) (Fig. 6A). As shown in Fig. 6B, primer pairs U2, S and M produced significantly higher signals from the anti-parafibromin immunoprecipitates than from those using control IgG. ChIP assays employing antibodies to other components of the PAF1 complex, Paf1 and Leo1, also gave strong specific signals with the U2, S and M primer pairs targeting CPEB1 (Fig. 6C–D). The specific ChIP signal was much stronger in the U2, U1, and D1 regions of CPEB1 than in the U3 region (Fig. 6E). Taken together, these data suggest that the PAF1 complex might be involved in both CPEB1 transcript initiation and elongation, consistent with PAF1 function at other gene loci (10–12, 27) and the observation above that Paf1 knockdown reduced CPEB1 transcript levels (Fig. 5E).
Figure 6. Chromatin immunoprecipitation demonstrates occupancy at CPEB1 but not CPEB3 by the PAF1 complex.
The physical association of endogenous parafibromin and other components of the PAF1 complex (including the Paf1 and Leo1 proteins) with the human CPEB1 promoter was examined by ChIP in HEK293 cells. (A) Schematic diagram showing the relative location of PCR primer sets employed in the ChIP assay along the human CPEB1 gene and flanking regions (not to scale). The negative numbers associated with U1-U3 indicate upstream position (in bp) of the primer sets relative to transcription start site, the positive number associated with D1 indicates downstream position (in bp) of the primer set relative to the end of the final gene exon. The neutral numbers associated with the early coding sequence (S) and middle coding sequence (M) represent the internal positions (in bp) of the primer sets 3’ to the transcription start site. (B–D): ChIP analysis using primer sets targeting upstream (U2), early coding (S) and middle coding (M) sequence of CPEB1 using either control IgG or antibodies against parafibromin (B), Paf1 (C) and Leo1 (D) proteins as shown. (E) ChIP analysis of parafibromin occupancy of regions upstream or the CPEB1 transcription start site or downstream of the end of the gene using the indicated primer sets. (F) ChIP analysis of the parafibromin, Paf1 and Leo1 occupancy of the human CPEB3 gene using the same ChIPed cell lysates employed in B-E. All experiments are representative of three or more independent biological repeats.
In contrast, none of the parafibromin, Paf1 and Leo1 specific antibodies yielded specific signal when interrogated with primers to the CPEB3 gene (Fig. 6F). These results suggest that parafibromin and the PAF1 complex play a direct regulatory role in the transcription of CPEB1 but not CPEB3, consistent with the failure of siRNA-mediated knockdown of Paf1 to affect CPEB3 transcript levels shown above (Fig. 5E).
Bioinformatic analysis of potential CPEB1 and hyx/HRPT2 target genes
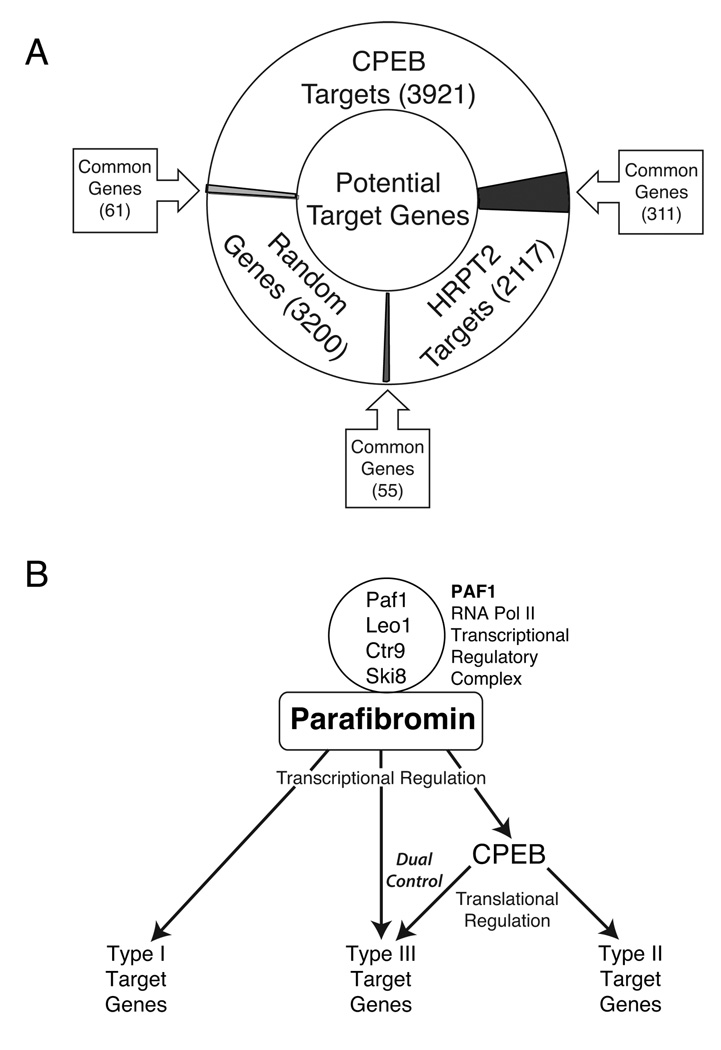
The regulation of CPEB1 gene transcription by HRPT2/CDC73 implies that parafibromin, apart from its transcriptional regulatory role as part of the PAF1 complex, might affect the translation of some genes indirectly through its regulation of CPEB1. In order to estimate the set of potential target genes regulated by both HRPT2/CDC73 and CPEB1 genes we performed a bioinformatic analysis. Whole genome microarray analysis was used to identify potential HRPT2/CDC73 target genes. In HEK293 cells there were 2117 genes whose transcription levels were either decreased or increased in response to RNA interference with, or cDNA transfection-mediated enhancement of, the expression of HRPT2/CDC73 (not shown). Potential CPEB1 target genes were identified by whole transcriptosome analysis using a computer program to identify transcripts with canonical CPE signals (U4–5A1–2U) located at an appropriate distance upstream of the poly(A) signal (AAUAAA) in their 3’ untranslated regions. This software analysis identified 3921 gene transcripts, a set that included known CPEB targets such as cyclin B1, c-Myc, and cdk1 (not shown). Comparison of the sets of potential HRPT2/CDC73 and CPEB1 target genes revealed 311 common genes, approximately 15% of the HRPT2/CDC73 targets (Supplemental Table I). To gauge the significance of this overlap, the sets of potential HRPT2/CDC73 and CPEB target genes were compared to a set of >3000 randomly selected genes. The overlap between the random set and the HRPT2/CDC73 and CPEB target sets was 2 and 3% respectively (Fig. 7A). The overlap of potential HRPT2/CDC73 and CPEB target gene sets is therefore highly significant (Fisher’s exact test, 2-tailed p value < 0.002, HRPT2/CPEB vs. HRPT2/Random; < 0.01, CPEB/HRPT2 vs. CPEB/Random).
Figure 7. Bioinformatic analysis of potential HRPT2 and CPEB targets suggests duel level of parafibromin gene control.
(A) Diagram showing the relationship of potential gene targets of HRPT2 and CPEB1 genes, and a set of randomly chosen genes. Potential HRPT2 targets (2117 genes) were identified by whole genome oligo microarray analysis comparing pools of transcript from HEK293 cells treated with either HRPT2-specific or scrambled control siRNA. Potential CPEB1 targets (3921 genes) were identified from the human genomic database based on the presence of a potential CPE consensus sequence in the 3’ untranslated region. To assess the specificity of the overlapping set of common HRPT2 and CPEB1 target genes, both pools of HRPT2 and CPEB1 potential target genes were also compared to a set of 3200 randomly-selcted human genes. An arrowed square box indicates the overlap identified by each of the three pairings (Fisher’s exact test, 2-tailed p value < 0.002, HRPT2/CPEB vs. HRPT2/Random; < 0.01, CPEB/HRPT2 vs. CPEB/Random). (B) Model illustrating three types of potential targets of parafibromin in association with PAF1 transcriptional regulatory complex: type I, regulated only transcriptionally (e.g. genes identified by whole genome oligo microarray analysis, not including the genes overlapping with CPEB1 targets); type II, regulated indirectly at the level of translation through CPEB1 (e.g. genes identified by CPE consensus sequence analysis, not including the genes overlapping with HRPT2 targets); and type III, regulated dually by transcription and indirectly by translational effects via CPEB1 (e.g. genes common to both HRPT2 and CPEB1 target gene pools). This model of dual regulation does not exclude the additional possibility of CPEB regulation by parafibromin involving direct physical complex formation, since the CPEB-binding scaffolding protein symplekin has been found in anti-parafibromin immunoprecipitates (18).
Discussion
Tumor suppressor genes generally induce programmed death or growth arrest (senescence) in cells malfunctioning because of genotoxic, oxidative, or nutritional stress, thus minimizing the deleterious effects of the cell on its neighbors (28, 29). The putative tumor suppressor gene HRPT2/CDC73 can induce growth arrest and apoptosis in vitro (14, 16) and in the present study we demonstrate an evolutionarily conserved pathway linking hyx/HRPT2 and orb2/CPEB and show that loss of function of either gene enhances starvation resistance and increases lifespan in a fly model.
The control of orb/CPEB gene expression by hyx/HRPT2 expands the potential mechanisms by which the tumor suppressor gene can control the expression of its targets (Fig. 7B). CPEB is a kinase-regulated RNA-binding protein component of the eIF-4E translation initiation complex that can regulate translation by either repressing or facilitating the cytoplasmic polyadenylation of a subset of 5’ capped mRNAs (24, 30). CPEB has been previously implicated in the regulation of cell proliferation and senescence (22), effects that may involve the translational regulation of tumor suppressor P53 expression (31). The bioinformatic analysis presented here suggests that the potential targets of the HRPT2/CDC73 include a subset of genes subject to both transcriptional and, through CPEB, translational control (Type III targets in Fig. 7B). Although the dual regulation of these putative target genes awaits experimental validation, it is tempting to think that HRPT2/CDC73 may exercise its tumor suppressor functions by utilizing a repertoire of distinct but reinforcing control mechanisms. The MYC protooncogene, for example, is repressed by parafibromin at both the transcriptional level and by a mechanism involving destabilization at the protein level (17). The potential for a third mechanism of control must now be considered since MYC is a target of CPEB (22) and was indeed identified as a potential target of both HRPT2/CDC73 and CPEB in the present bioinformatic analysis (Supplemental Table I).
Although the linkage between hyx/parafibromin and orb/CPEB demonstrated here is at the transcriptional level other mechanisms must be considered. In budding yeast the Paf1 complex including the parafibromin homolog Cdc73p can directly interact with the 3’-mRNA processing cleavage and polyadenylation factor Cft1 (32). In cultured human cells, multiple subunits of the cleavage and polyadenylation specificity factor (CPSF) as well as the cleavage stimulation factor were recently identified by mass spectroscopy in anti-parafibromin immunoprecipitates (18). Since these immunoprecipitates also contained symplekin (18), a putative scaffolding protein that binds both CPEB and CPSF and is required for CPEB-mediated polyadenylation (33), it is possible that a physical regulatory complex containing parafibromin and CPEB may also exist.
The rescue phenotype of the lobe/hyx double heterozygote we report may provide fresh insight into the earlier observation that hyx/parafibromin binds directly to armadillo/β-catenin and facilitates Wg/Wnt signaling (13). Previous studies of the developing Drosophila eye showed that lobe was required for early cell survival, and that loss of lobe function was associated with the induction of cell death and upregulation of Wg signalling (20) and indeed our studies confirm a marked increase in the number of apoptotic nuclei in the eye imaginal discs of Lsi/+ heterozygote larvae (Fig. 3A, C). Singh et al furthermore found that blockade of Wg signaling could rescue the lobe loss-of-function phenotype (20). In this light it is tempting to speculate that the rescue of lobe loss-of-function in lobe/hyx double heterozygotes may reflect the requirement of hyx/parafibromin for nuclear transduction of the Wg/Wnt signal (13).
Two lines of evidence presented here suggest that the tumor suppressor function of hyx/parafibromin may result in part from an involvement in nutritional sensing pathways. First, as discussed above, it was found that hyx/HRPT2 heterozygosity could rescue the loss of function phenotype of lobe, the fly homolog of PRAS40. PRAS40 is a raptor-interacting protein and target of the mTOR kinase that can inhibit cell growth under conditions of nutritional stress (see (34) for recent review). Secondly we observed that heterozygous hyx/HRPT2 loss of function enhanced fly longevity and imparted resistance to starvation. Orb2/CPEB heterozygosity had a similar effect on longevity and nutritional stress resistance. At the cellular level it is clear that heightened resistance to nutritional stress resulting from hyx/HRPT2 or orb/CPEB loss of function would enhance tumorigenesis by promoting the survival of rapidly dividing and hypermetabolic tumor cells as levels of available nutrients decline. Future work will help resolve the critical interactions linking parafibromin to nutrient sensing machinery and other pathways involved in tumorigenesis.
Materials and methods
Fly stocks
The enhancer trapped fly lines from the Japanese NP Consortium Gal4 Enhancer Trap Insertion Database (GETDB) were obtained from Drosophila Genetic Resource Center (DGRC), Kyoto Institute of Technology, Kyoto, Japan. The fly line bearing the hypomorphic allele, hyxEY6898, which contains a P-element (P[EPgy2]) insertion located 36 bp upstream of the hyx translational start site in the 5’ untranslated transcript region was originally obtained from the Gene Disruption Project (GDP), Baylor College of Medicine, Texas, but is now available from the Bloomington Drosophila Stock Center at Indiana University (Stock No. 16768). The hyxdEY2/+ fly was created via imprecise excision of P[EPgy2] by crossing hyxEY6898 with the transposase expressing fly line, Pi(Δ2–3)k To create the doubly heterozygous mutant fly line (Lsi/cyo; hyxEY6898/Tm3), standard genotype synthesis methodology employing both the second and third chromosome balancers (Cyo/Sco and Tm6/Tm3) was used (19). All the other fly lines used were obtained from the Bloomington Drosophila Stock Center at Indiana University.
Morphogenesis, histology and scanning electron microscopy
Fly morphogenesis during development was characterized using stereomicroscopy. Wild type flies and those with different genetic mutations were synchronized developmentally by collecting embryos every hour and examined morphologically at different developmental stages by stereomicroscopy (Zeiss Stemi 2000-C). Detailed structures of fly eyes were further studied using variable pressure vacuum scanning electron microscopy (Hitachi S-3400N VP SEM). Flies were first fixed in 4% paraformaldehyde (Electron Microscopy Sciences) overnight and then maintained in 100% ethanol. Immediately before use, fly heads were removed and examined under appropriate vacuum pressure so that the fly eyes maintained their shapes for 10–15 minutes before deformation. Paraffin-embedded sections of fly larvae were prepared by Histoserv, Inc (Germantown, MD) and examined by standard light microscopy.
Single larva genotyping
In order to create Lsi/+; hyxEY6898/orb2BG02373 triply heterozygous larvae, Lsi/Lsi; hyxEY6898/Tm3 heterozygotes were crossed with orb2BG02373/orb2BG02373 flies. After the mouthpart was removed from each of the resulting third instar larvae, the remainder of the body was put individually into a single well of a 96 well plate containing 50 µl of DirectPCR lysis reagent (Viagen, Cat# 102-T) and incubated at 85 °C for 45 min. PCR reactions were performed using 2 µl of the supernatant and the Brilliant® II SYBR® Green QPCR Master Mix kit (Agilent Technologies) in a 20 µl of total reaction volume. The PCR cycling conditions used were: 95 °C 10’, followed by 40 cycles of 95 °C X 30’’, 55 °C X 1 min, and 72 °C X 1 min. The primer pairs used to detect the wild-type hyx gene were: 5’-GAGAAGCGATGCACTCTCTATG-3’and 5’-GCTACGCACTTTGTAATCCGCGAAAG-3’; and for the mutant hyx gene were: 5’-CAATCATATCGCTGTCTCACTCA-3’ and 5’-GCTACGCACTTTGTAATCCGCGAAAG-3’.
Larval eye disc apoptosis and cell proliferation analysis
The ApopTag Red In Situ Apoptosis Detection Kit (Millipore) was used for TUNEL analysis. Eye discs from third instar larvae were dissected in PBS and fixed in 4% paraformaldehyde for 20 min at RT, washed 3 x in PBTween (5 min ea), post-fixed in pre-cooled 2:1 EtOH:PBS for 5 min at −20° C, and washed 2 X in PBTween (5 min ea). Eye discs were then incubated in 10mM sodium citrate (pH 6.0) for 30 min at 70° C, and rinsed in pure dH2O for 10 min at RT. Tissue was then incubated in working strength TdT Enzyme (prepared according to manufacturer’s instructions) for 1h at 37° C, incubated in 1X Stop/Wash solution for 10 min at RT, and washed 3 x in PBTween (1 min ea) at RT. Tissue was then incubated in ApopTag anti-digoxigenin with rhodamine for 30 min at RT, protected from the light. Eye discs were then washed 4 x in PBTween (2 min ea) at RT, mounted in Vectashield Mounting Solution with DAPI, and analyzed using fluorescence microscopy. For the detection of cellular proliferation in third instar eye imaginal discs the Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen) was used. Late second instar larvae were collected and fed a 200 µl solution of 20 µM 5-ethynyl-2’-deoxyuridine (EdU) per g of food for 24 hr. Eye discs were then dissected in PBS and fixed in 4% paraformaldehyde for 20min at RT. Eye discs were washed 2 x with PBS, (2 min ea) and incubated in 0.5% Triton X-100 for 30 min at RT. A Click-iT reaction cocktail (containing Alexa 488 azide for the detection of EdU incorporation) was prepared according to the manufacturer’s instructions. Eye discs were incubated in the Click-iT reaction cocktail for 30 min at RT, protected from the light, and then rinsed briefly in Click-iT reaction rinse buffer. Eye discs were then mounted in Vectashield Mounting Solution with DAPI, and analyzed using fluorescence microscopy. Clusters of EdU-positive cells anterior to the morphogenetic furrow were counted in wild-type or triple heterozygote larval eye discs if their brightness exceeded that of the average level in the antennal portion of the imaginal disc, used as an internal reference.
mRNA quantification
Gene expression levels were estimated based on transcript abundance as measured by quantitative RT-PCR and oligo microarray analysis. Quantitative RT-PCR was performed with one step quantitative RT-PCR master mix (Agilent Technologies) using a Stratagene MX 3000P real time PCR machine and analyzed using the accompanying software. For each experiment, β-actin was used for normalization. For every run, standard curves from 4–5 points of 1:4 serial dilutions of both β-actin and the target gene were performed to minimize differences between runs. Each reaction was conducted in triplicate and 3–9 biological samples prepared independently were used in data analysis. The Prism software version 5.0b (GraphPad Software, Inc.) was used for graphing of the analyzed data set. Microarray analysis was performed using the Affymetrix whole genome DNA array. Total RNA was prepared from both treated and control samples using QIAGEN RNeasy kit. RNA probe preparation, hybridization and primary data analysis were performed by the NIH/NIDDK microarray core facility.
Genetic screening for hyx/HRPT2 gene modifiers
The first phase of genetic screening was conducted using the hyx/HRPT2 gene P element mutation line EY6898/TM3 and approximately 400 target fly stock lines obtained from either the Bloomington or Japanese GETDB fly stock centers. The target fly lines chosen for screening contained mutant alleles of fly genes homologous to human genes implicated in oncogenic, tumor suppressor or stress resistance pathways. Crosses that produced offspring with altered phenotypes were recorded. The second phase of genetic screening was performed using a triple gene interaction strategy. The doubly heterozygous mutant Lsi/cyo; hyxEY6898/Tm3 (generated by genotype synthesis as described above) was used to re-screen the 400 target fly lines. Crosses that produced offspring with eye phenotypes different from the parents were recorded. To determine if the novel phenotype resulted from the interaction of the target gene with lobe gene or with hyx/HRPT2 gene, lobe/ target gene double heterozygotes and hyx/HRPT2/ target gene double heterozygotes were also examined for eye phenotype changes.
Antibodies, mammalian cDNA expression constructs, and cell culture
Antibodies used included goat anti-CPEB (K-16) antibody (sc-48983, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-human parafibromin antibody GRAPE-2 (14), mouse anti-AU5 monoclonal (MMS-135R, Covance Research Products, Denver, PA), mouse anti-β-actin monoclonal (A5316, Sigma, St. Louis, MO), rabbit polyclonal anti-Leo1 (A300-175A, Bethyl Laboratories, Inc., Montgomery, TX) and rabbit polyclonal anti-Paf1 (A300-172A, Bethyl Labs). Secondary antibodies utilized in immunoblots were Cy3-conjugated donkey anti-mouse IgG (715-165-150, Jackson ImmunoResearch Labs, West Grove, PA) and IR secondary antibodies (anti-rabbit IR 800 and anti-mouse Red and Green) from LI-COR Bioscience. RNA interference methodology and the sequences of parafibromin- and Paf1-directed siRNAs were previously described (14, 17). Complementary cDNA for AU5-epitope N-terminally tagged human parafibromin was previously described (15). AU5-tagged human parafibromin cDNA with silent base changes introduced to impair interaction with siRNA construct siPfb-1 was prepared using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene) and a mutagenic primer pair consisting of the sense primer 5’-CAG ACT GAA CAG ATT AGG agc cTa TCT GAA GCT ATG TCA GTG-3’ and its reverse complement (bases in lower case represent silent changes). The coding region of the siRNA-resistant cDNA was confirmed by DNA sequencing. Human embryonic kidney HEK 293 and HEK-293FT cells were grown in 75 cm2 flasks in DMEM supplemented with 10% fetal bovine serum, 4mM L-glutamine and penicillin/streptomycin at 37° C and 5 % CO2. Empty vector or expression plasmid were transfected using Lipofectamine 2000 (Invitrogen).
Immunoblotting, chemiluminescence and infrared imaging
Cell lysates were boiled with equal volume of Laemmli’s 2X gel loading buffer and the hot solution was loaded onto 4–20% Tris-Glycine SDS-PAGE gels (Invitrogen) to separate the proteins, followed by transfer of the proteins on to 0.45-micron nitrocellulose membrane. Membranes were blocked with TBS or PBS (pH 7.4) containing 0.1% Tween 20 and 5% nonfat dry milk (blocking buffer) and incubated overnight with primary antibodies in the same buffer. The membranes were then washed seven times for 5 minutes each with the above buffer without milk, followed by a 2-hour incubation in blocking buffer including appropriate horseradish peroxidase-conjugated secondary antibodies. Membranes were then washed as above, and the proteins detected by chemiluminescence on X-ray film using Super Signal West Dura Extended Duration Substrate (Pierce). For infrared (IR) imaging, IR-labeled secondary antibodies (dilution 1: 20,000) were used for detecting the protein signals in conjunction with the Odyssey infrared imaging system (LI-COR, Bioscience). Blocking and washing buffers used were same as described above, however the incubation time with the secondary antibody was half hour- one hour protected from light. For the quantification of the intensity of the protein bands membranes were dually probed, with the β-actin used as a loading control.
Lifespan and stress resistance
The lifespan and stress resistance of heterozygous hyx/HRPT2 flies obtained by crossing hyx/Tm3 with w1118 flies were examined as compared to wild type (w1118) flies. Newly eclosed flies within a period of 24 hrs were collected and aged for 5 days on standard corn meal agar fly medium. Male and female flies were then separated for testing. For lifespan test, 20 flies were transferred to a vial containing 10 ml of culture medium maintained at 25 °C. Flies were transferred to fresh medium every 3 days and the number of dead flies were recorded. The survival rate per vial at each recording time was calculated by the formula: (1- [dead flies/total flies]) x 100%. Ten or more vials were used for each experiment and 3 or more independent experiments were conducted for each fly line. For data analysis, each vial was treated as a data point and analyzed by Prism software version 5.0b (GraphPad Software, Inc.). For oxidative and starvation stress tests, flies with desired genotypes eclosed within 24 hrs were collected. After ageing for 5 days flies were separated by gender and placed into vials containing Whatman paper discs (d=2.3 cm) soaked either in 350 µl of 5% sucrose solutions with or without paraquat for paraquat treatments or in H2O for starvation tests. Five to ten vials (10 flies per vial) were used for each test and at least 3 independent experiments were performed. Dead flies were recorded daily and statistical analysis was performed using the Prism software as described above.
Chromatin immunoprecipitation assay
ChIP assay kit from Upstate (Cat. No. 17–295) was used in the analysis of HEK293 cells following the manufacturer’s instructions except that the QIAquick PCR purification kit (Qiagen, Cat. No. 28104) was used for DNA purification. Purified DNA was used to amplify CPEB1 and its flanking sequences using the following primer pairs: U1 (upstream 753 bp, ATCAAGCAAAGGCAGAGAGGGA, AAACAGACCCGACAACTGCCAA); U2 (upstream 1430 bp AGCTTCTTTGGGTTGCTGAGGT, TCCTGGAGAAAGCATGGCTCAA); U3 (upstream 2677 bp, AAACAGCCTTTGAGCCCAGCTA, TCCTGCAGAAGCACTGAACACT ); S (early coding 19210 bp, TTTCACATTGAGCAGGCCGAG, ACTGTGCCTGCTTCTCCTTACA); M ( middle coding 19808 bp, GGATTTCTCCAAGGTCCATGTC, TCCATGAAAGCCATCATGCCCA); D1(downstream 1105 bp, ATGTTGCTCAGGCTGGTCTCAA, TGGCTCACGCTTACAATCAGCA). The primer set, GCGCTCGTTTTGTGCAGCTTC and GTGCCCTGGCACTCATCACAC, was used to amplify the CPEB3 gene.
Bioinformatic analysis
CPEB binds specifically to a cytoplasmic polyadenylation element (CPE) located upstream, mostly within 100 bases, of the hexanucleotide poly(A) signal (AAUAAA) sequence at the 3’ UTR of mRNA to control polyadenylation of mRNAs (30). The consensus CPE has the general form: U4–5A1–2U. A PERL script was developed to identify every gene in the human RNA database that contains the conserved pattern: U4–5A1–2U N1–100 AAUAAA where N can be any nucleotide. For quality control of the PERL script-identified genes, we randomly checked about 10% of the genes selected by the program. 100% of the individually examined genes contained the desired consensus sequence pattern. The bioinformatically-selected putative CPEB target genes were compared with potential parafibromin target genes selected by whole genome oligo microarray analysis to identify the common targets of both proteins. To evaluate the specificity of the shared targets identified by this method, each set of the targets was compared independently with a set of 3200 genes randomly selected from the NCBI Reference Sequence (RefSeq) database using Insightful-Miner (TIBCO Software, Inc.).
Supplementary Material
Acknowledgements
We are grateful to Sunita Agarwal and Stephen Marx for encouragement and helpful discussions. This research was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Abbreviations
- CPEB
cytoplasmic polyadenylation element binding protein
- HPT-JT
hyperparathyroidism-jaw tumor syndrome
- ChIP
Chromatin immunoprecipitation
Footnotes
Conflict of Interest
None of the authors has a conflict of interest that could be perceived as prejudicing the impartiality of the research reported herein.
REFERENCES
- 1.Jackson CE, Norum RA, Boyd SB, Talpos GB, Wilson SD, Taggart RT, et al. Hereditary hyperparathyroidism and multiple ossifying jaw fibromas: a clinically and genetically distinct syndrome. Surgery. 1990;108:1006–1012. [PubMed] [Google Scholar]
- 2.Mallette LE, Malini S, Rappaport MP, Kirkland JL. Familial cystic parathyroid adenomatosis. Ann Intern Med. 1987;107:54–60. doi: 10.7326/0003-4819-107-1-54. [DOI] [PubMed] [Google Scholar]
- 3.Teh BT, Farnebo F, Kristoffersson U, Sundelin B, Cardinal J, Axelson R, et al. Autosomal dominant primary hyperparathyroidism and jaw tumor syndrome associated with renal hamartomas and cystic kidney disease: linkage to 1q21-q32 and loss of the wild type allele in renal hamartomas. J Clin Endocrinol Metab. 1996;81:4204–4211. doi: 10.1210/jcem.81.12.8954016. [DOI] [PubMed] [Google Scholar]
- 4.Teh BT, Farnebo F, Twigg S, Höög A, Kytölä S, Korpi-Hyövälti E, et al. Familial isolated hyperparathyroidism maps to the hyperparathyroidism-jaw tumor locus in 1q21-q32 in a subset of families. J Clin Endocrinol Metab. 1998;83:2114–2120. doi: 10.1210/jcem.83.6.4896. [DOI] [PubMed] [Google Scholar]
- 5.Simonds WF, James-Newton LA, Agarwal SK, Yang B, Skarulis MC, Hendy GN, et al. Familial isolated hyperparathyroidism: Clinical and genetic characteristics of thirty-six kindreds. Medicine (Baltimore) 2002;81:1–26. doi: 10.1097/00005792-200201000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Simonds WF, Robbins CM, Agarwal SK, Hendy GN, Carpten JD, Marx SJ. Familial isolated hyperparathyroidism is rarely caused by germline mutation in HRPT2, the gene for the hyperparathyroidism-jaw tumor syndrome. J Clin Endocrinol Metab. 2004 Jan;89(1):96–102. doi: 10.1210/jc.2003-030675. [DOI] [PubMed] [Google Scholar]
- 7.Carpten JD, Robbins CM, Villablanca A, Forsberg L, Presciuttini S, Bailey-Wilson J, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet. 2002 Dec;32(4):676–680. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- 8.Shattuck TM, Valimaki S, Obara T, Gaz RD, Clark OH, Shoback D, et al. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N Engl J Med. 2003 Oct 30;349(18):1722–1729. doi: 10.1056/NEJMoa031237. [DOI] [PubMed] [Google Scholar]
- 9.Cetani F, Pardi E, Borsari S, Viacava P, Dipollina G, Cianferotti L, et al. Genetic analyses of the HRPT2 gene in primary hyperparathyroidism: germline and somatic mutations in familial and sporadic parathyroid tumors. J Clin Endocrinol Metab. 2004 Nov;89(11):5583–5591. doi: 10.1210/jc.2004-0294. [DOI] [PubMed] [Google Scholar]
- 10.Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, et al. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005 Jan;25(2):612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, et al. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005 Jun;25(12):5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, et al. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005 Jul 15;19(14):1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006 Apr 21;125(2):327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- 14.Lin L, Czapiga M, Nini L, Zhang JH, Simonds WF. Nuclear localization of the parafibromin tumor suppressor protein implicated in the hyperparathyroidism-jaw tumor syndrome enhances its proapoptotic function. Mol Cancer Res. 2007 Feb;5(2):183–193. doi: 10.1158/1541-7786.MCR-06-0129. [DOI] [PubMed] [Google Scholar]
- 15.Woodard GE, Lin L, Zhang JH, Agarwal SK, Marx SJ, Simonds WF. Parafibromin, product of the hyperparathyroidism-jaw tumor syndrome gene HRPT2, regulates cyclin D1/PRAD1 expression. Oncogene. 2005 Feb 10;24(7):1272–1276. doi: 10.1038/sj.onc.1208274. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, Kong D, Tan MH, Pappas DL, Jr, Wang PF, Chen J, et al. Parafibromin inhibits cancer cell growth and causes G1 phase arrest. Biochem Biophys Res Commun. 2006 Nov 10;350(1):17–24. doi: 10.1016/j.bbrc.2006.08.169. [DOI] [PubMed] [Google Scholar]
- 17.Lin L, Zhang JH, Panicker LM, Simonds WF. The parafibromin tumor suppressor protein inhibits cell proliferation by repression of the c-myc proto-oncogene. Proc Natl Acad Sci U S A. 2008 Nov 11;105(45):17420–17425. doi: 10.1073/pnas.0710725105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3’ mRNA processing factors. Proc Natl Acad Sci U S A. 2009 Jan 20;106(3):755–760. doi: 10.1073/pnas.0812023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenspan RJ. Fly Pushing: The Theory and Practice of Drosophila Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. Chapter 4. Synthesizing specific genotypes; pp. 63–86. [Google Scholar]
- 20.Singh A, Shi X, Choi KW. Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development (Cambridge, England) 2006 Dec;133(23):4771–4781. doi: 10.1242/dev.02686. [DOI] [PubMed]
- 21.Chern JJ, Choi KW. Lobe mediates Notch signaling to control domain-specific growth in the Drosophila eye disc. Development (Cambridge, England) 2002 Sep;129(17):4005–4013. doi: 10.1242/dev.129.17.4005. [DOI] [PubMed] [Google Scholar]
- 22.Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. Control of cellular senescence by CPEB. Genes Dev. 2006 Oct 1;20(19):2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasko P. Gene regulation at the RNA layer: RNA binding proteins in intercellular signaling networks. Sci STKE. 2003 Apr 22;2003(179):RE6. doi: 10.1126/stke.2003.179.re6. [DOI] [PubMed] [Google Scholar]
- 24.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007 June;32(6):279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Reifegerste R, Moses K. Genetics of epithelial polarity and pattern in the Drosophila retina. BioEssays. 1999 Apr;21(4):275–285. doi: 10.1002/(SICI)1521-1878(199904)21:4<275::AID-BIES3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007 Sep;8(9):729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 27.Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. MolCellBiol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005 Feb 25;120(4):513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Campisi J. Aging, tumor suppression and cancer: high wire-act! Mech Ageing Dev. 2005 Jan;126(1):51–58. doi: 10.1016/j.mad.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 30.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol. 2001 Jul;2(7):521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 31.Burns DM, Richter JD. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008 Dec 15;22(24):3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nordick K, Hoffman MG, Betz JL, Jaehning JA. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryot Cell. 2008 Jul;7(7):1158–1167. doi: 10.1128/EC.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004 Nov 24;119(5):641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 34.Dunlop EA, Tee AR. Mammalian target of rapamycin complex 1: signalling inputs, substrates and feedback mechanisms. Cell Signal. 2009 Jun;21(6):827–835. doi: 10.1016/j.cellsig.2009.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.