Abstract
We have used P element deletion derivatives at defined locations in the Drosophila genome to construct a 100-kb extended P element more than twice the size of any previously available. We demonstrate that this prototypical extended P element is capable of transposition to new sites in the genome. The structural and functional integrity of a transposed extended P element was confirmed using molecular, genetic, and cytogenetic criteria. This is the first method shown to be capable of producing large, unlinked transpositional duplications in Drosophila. The ability to produce functional transposable elements from half-elements is novel and has many potential applications for the functional analysis of complex genomes.
Functional analysis of complex eukaryotic genomes requires continued development of experimental methods for mapping and assessing gene function on a genome-wide scale. A well-established model system for the development of such methods has been the fruit fly Drosophila melanogaster. In Drosophila, two of the most valuable tools for genetic analysis of the genome, chromosome rearrangements and transposable elements, complement one another. Chromosome rearrangements, particularly deficiencies and duplications, are useful for genetic mapping within large chromosomal regions, whereas transposable elements have been used primarily for the identification and analysis of small genomic regions and single genes. In D. melanogaster, the many chromosomal deficiencies that have been isolated and cytologically mapped provide an invaluable resource for the rapid mapping of mutations and for a variety of genetic screens (Lindsley and Zimm 1992; Smith et al. 1993; Bilder and Scott 1995; Staehling-Hampton et al. 1999). Their utility is limited, however, because the continuity and endpoints of the deletions are not well defined and because reductions in viability and other phenotypes often associated with multigenic deletions can complicate the interpretation of complementation tests used for genetic mapping.
Most spontaneous mutations in Drosophila result from the insertion of transposable elements (Zachar and Bingham 1982), which have been indispensable in the molecular genetic analysis of the Drosophila genome. The discovery of the P transposable element provided the means for the rapid identification and analysis of Drosophila genes through the development of P-element-mediated germ-line transformation and methods for P-element insertional mutagenesis (Engels 1996). Most recently, genome-scale P-element insertional mutagenesis has resulted in the identification of lethal mutations for an estimated 25% of the essential loci in Drosophila (Spradling et al. 1999).
Transposition and excision of P elements requires specific cis-acting DNA sequences found at the 5′ and 3′ ends of the transposon as well as P transposase and host factors (O'Hare and Rubin 1983; Karess and Rubin 1984; Rio and Rubin 1988; Beall and Rio 1996). Although the two P-element ends share common sequence motifs, they are not functionally equivalent, and functional P elements require one 5′ end and one 3′ end (Mullins et al. 1989). The particular DNA sequences placed between the P-element ends have little effect on the element's ability to undergo excision and transposition, but the rates of both germ-line transformation and in vivo transposition are inversely correlated with total P-element size (Spradling 1986). The largest P elements introduced into the Drosophila genome have been based on cosmid vectors (Haenlin et al. 1985), setting an arbitrary upper size limit of about 40 kb for determining the relationship between transposon size and transposition frequency. Larger P elements have been constructed using bacterial recombination (O'Connor et al. 1989), but germ-line transformants were not reported, perhaps because P-element-mediated germ-line transformation requires the isolation and injection of high-quality, closed circular DNA (Engels 1996). Thus, while cosmid-based P elements can undergo in vivo transposition to new sites in the genome at appreciable frequencies, the transposition rates for larger P elements are unknown.
To overcome the size limitations associated with P-element-mediated germ-line transformation, we constructed larger P elements by in vivo manipulation of pre-existing P-element insertions. This in vivo experimental approach is based on identifying P-element deletion derivatives that have lost one or the other end (5′ or 3′) of the P element, which we refer to as half-P elements. By placing properly orientated half-P elements on the same chromosome, we create extended P elements, in which one half-P element, containing a functional 5′ end, is physically linked to another half-P element containing a functional 3′ end. The resulting extended P element is composed of two compatible half-P elements separated by a large region of genomic DNA.
Here we report the successful transposition of a 100-kb prototypical extended P element from the third chromosome to the X chromosome, demonstrating the feasibility of constructing functional extended P elements from half-P elements. Such transposition events create transgenomic P elements that have a variety of potential applications for functional genomics, including acting as unlinked transpositional duplications and as large P elements capable of further manipulation. Using a modification of this approach, we can now construct a large set of extended P elements in order to evaluate the effects of size and position on transposition and excision frequencies. Our ultimate goal is the creation of a set of transgenomic P elements that collectively span most of the Drosophila genome. We discuss the potential of this technology for the manipulation and analysis of complex genomes.
RESULTS
We have employed a combination of genetic screens and molecular analysis to create, by stepwise construction, a large extended P element composed of two compatible half-P elements separated by a large region of genomic DNA. The orientation and genomic insertion sites for the two P elements used to produce the prototypical 100-kb extended P element, ExtP1, are summarized in Figure 1. The three-step construction of ExtP1 is presented schematically in Figure 2A, and the genetic crosses carried out for each of the three steps, as well as for transposition of ExtP1 (step 4), are diagrammed in Figure 2B.
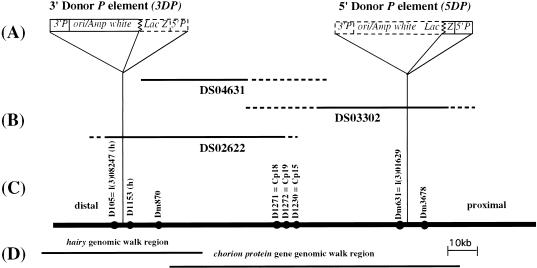
Figure 1.
The genomic region encompassed by the extended P element. (A) The structure of the two P elements used to construct the extended P element is shown. The two closely linked P elements (3DP and 5DP) used for the construction of the extended P element contain the bacterial orgin of replication (ori), the bacterial ampicillin resistance gene (amp) and LacZ gene, and the Drosophila white gene, as well as the 5′ and 3′P element ends 3′P and 5′P (Haenlin et al. 1985). Dashed lines outline the regions of the P elements deleted during the production of half-P-element derivatives. (B) Position of P1 genomic phage in the 66D region used in this study. Uncertainty in the endpoints of P1 genomic clones are shown in dashed lines. (C) Genomic DNA in the 66D region (shown as a solid line) including chromosomal orientation (distal and proximal to the centromere) and the location of sequence tagged sites (sts) in the region, shown as solid circles. (D) Extent of two overlapping genomic λ phage walks through the region, used to characterize the extended P element further, are shown (Spradline 1981; Ish-Horowitz et al. 1985).
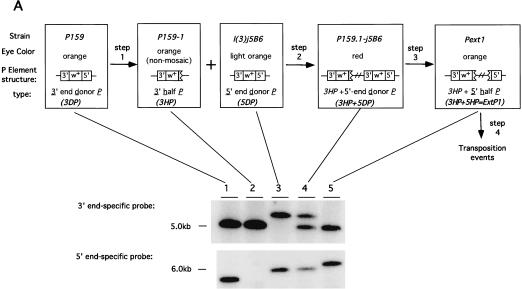
Figure 2.
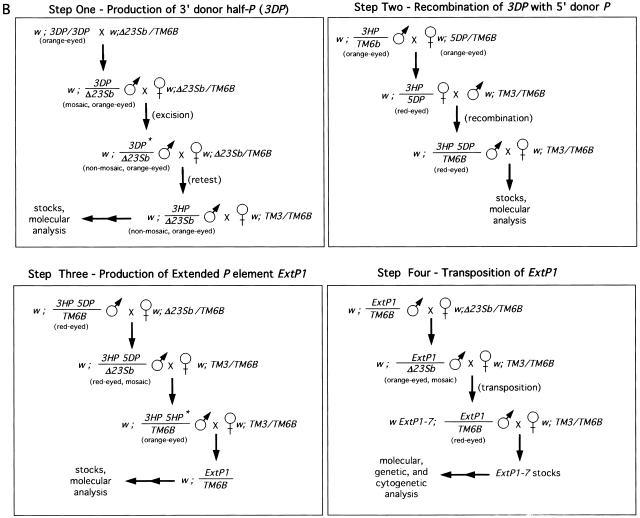
Construction and transposition of the prototypical extended P element, ExtP1. (A) Shown on the top are the three steps used to construct ExtP1. Each rectangular box shows the name of the strains utilized for each step followed by the eye color observed, structure of the P element, and type of P element. Southern blot analysis of the structure of each element, determined using 3′- and 5′-end-specific probes, is shown below. (B) Representative crosses carried out to perform each step shown in A. Abbreviations for balancer chromosomes used are: TM3 = TM3, SB ry e and TM6B = TM6B, Tb Hu e. All other abbreviations are given in A and in Results.
In Vivo Construction of the Extended P Element, ExtP1, on Chromosome Arm 3L
We first identified half-P elements by means of a phenotypic screen for nonmosaic, orange-eyed flies that retained a functional white gene but could no longer undergo detectable somatic excision and transposition in the presence of somatically expressed P transposase, shown as step 1 in Figure 2. From this screen we recovered nonmosaic progeny at frequencies ranging from 0.1% to 1%. Further genetic and molecular analysis of the resulting strains led to the identification of the half-P deletion-derivative strain, P159-1, used for the construction of the prototype extended P element (Fig. 1; and see Methods). This strain contains a half-P element, 3′P{w+}159.1, hereafter referred to as 3HP, inserted in the upstream region of the hairy gene at cytological position 66D10 (see Methods for element nomenclature). The 3HP element retains the 3′ end of the P element, the bacterial vector sequences, and the white gene, but has lost the 5′ end of the P element (Fig. 2A, lanes 1,2). Using P element end-specific probes (see Methods), we confirmed that loss of mosaicism in the P159-1 strain was correlated with the loss of cis-acting sequences at the 5′ end of the P element (see Fig. 2). The orientation of both the 3DP and 3HP was determined by sequence and hybridization of plasmid rescued sequences (Fig. 1).
Next, to identify a second P element insertion compatible with the 3HP element, which has its 3′ end distal to the centromere, we sought a proximal P element insertion in the same orientation as the 3HP element. From among >20 strains believed to contain proximally located P elements, we identified the l(3)j5B6 strain as having a properly positioned and oriented P element. This strain contains a lethal P element insertion, P{lacW}-j5B6, hereafter referred to as 5DP (for 5′ donor P element; Fig. 1).
Half-P elements retaining the internal white gene as a marker were obtained at only low frequencies from the 3DP element (see Methods). This observation, as well as the identification of other P-element fusion derivatives containing multiple ends (data not shown) suggested that the half-P element derivatives retaining the white gene were produced as a result of multistep processes rather than by assymetric gap repair following P element excision. In contrast, smaller P-element deletion derivatives that have lost the internal marker gene as well as one of the ends occur more frequently (Svoboda et al. 1995). We therefore identified putative excision derivatives of the 5DP element under conditions that would allow us to identify half-P derivatives without a requirement for maintenance of the internal white marker gene. To do this, we placed the full-length 5DP element into the cis configuration with the 3HP element by meiotic recombination, shown as step 2 in Figure 2. The resulting strain, denoted P159.1-j5B6, has red eye color as a result of the presence of two copies of the white gene. This strain contains two 3′ ends, one from the 3HP element and one from the 5DP element, as well as a single 5′ end from the 5DP element (Fig. 2A, lane 4). This strain was then used in a phenotypic screen for orange-eyed flies expected to have lost most or all of the 5DP element (including the white gene) while retaining the white gene from the 3HP element, shown as step 3 in Figure 2. Molecular analysis of >100 independent orange-eyed progeny led to the identification of the 5′ half-P element 5′P{w−}5B6-54, hereafter referred to as 5HP. The 5HP element retained 921 bp of the original 5DP element, including all the 5′ sequences required for transposition (Spradling et al. 1999) and a small segment of the LacZ gene, fused to the 31-bp inverted repeat retained from the 3′ end of the 5DP element. The rest of the original 5DP element had been deleted, including most of the 3′ cis-acting DNA sequences required for transposition (Fig. 2, lane 5). Because the 5′ end provided by the 5DP element was produced on the same chromosome containing the compatible 3HP derivative, the resulting strain, denoted Pext1, contains a prototypical extended P element, P{Ext, w+}1, hereafter referred to as ExtP1. The entire genomic region encompassed by ExtP1 is known to be ∼100 kb in length, on the basis of two overlapping chromosome walks (Spradling 1981; Ish-Horowicz 1985) and available genomic DNA sequence.
Excision and Transposition of the ExtP1 Extended P Element
To determine whether the extended P element was capable of excision and transposition to new sites in the genome, we mobilized ExtP1 using standard genetic methods (Fig. 2B, step 4). Eye-color mosaicism in the presence of the P transposase source provided the first evidence that ExtP1 can undergo excision and transposition (data not shown). Thirty-five excision events were isolated on the basis of loss of the white gene among ∼1.2 × 103 progeny, a rate of ∼0.3%. In a similar screen we identified putative transposition events as red-eyed progeny containing two copies of the white gene. From segregation analysis of 80 such red-eyed flies, we identified two interchromosomal transposition events, providing a rough estimate of 0.01% for the rate of interchromosomal transposition. Previous estimates of these rates for marked P elements have generally fallen between 1% and 20%, and larger elements have yielded significantly lower rates (Engels 1996; Spradling 1986).
Cytological Integrity of ExtP1-7 on the X Chromosome
In one of the strains bearing a putative interchromosomal transposition event, orange eye color segregated with the X chromosome. We examined the structural integrity of this X-linked extended P element, denoted ExtP1-7, at the cytogenetic level using high-resolution fluorescence in situ hybridization (FISH) to visualize the position, orientation, and arrangement of the transposed sequences. Figure 3 shows images taken from polytene chromosome squashes after triple fluorescent labeling to reveal the chromosomes (DAPI, blue), half-Ps (green), and internal genomic DNA (red), respectively. Hybridization with a probe of labeled P-element vector sequences defines the cytological position and boundaries of the extended P element, and hybridization with a probe of labeled P1 phage clone DS04631 (Fig. 1) confirmed that genomic DNA from the third chromosome was in fact present on the X chromosome (Fig. 3, panels A–D).
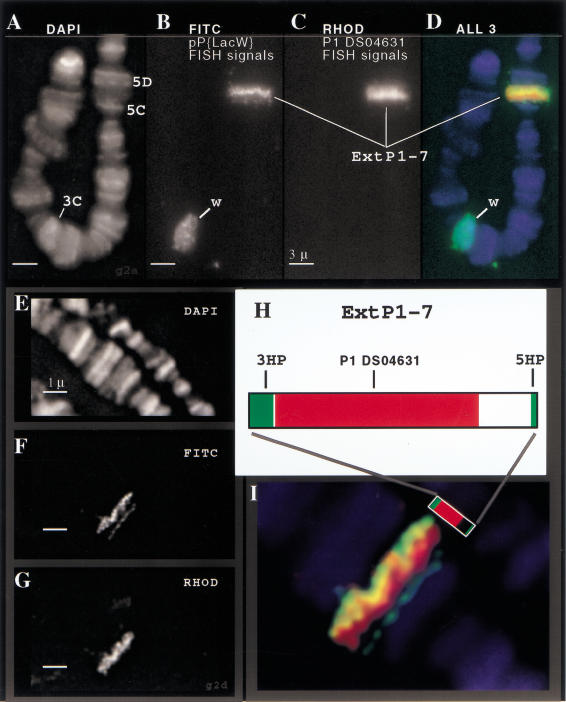
Figure 3.
Cytogenetic fine structure FISH analysis of ExtP1-7. Direct-labeled FISH probes were used to visualize the extended P element, ExtP1-7, on the X chromosome. Three-color staining allowed simultaneous detection of chromatin (DAPI images, panels A,E), the boundaries of the extended P elements (pP{LacW} FISH signals—FITC channel, panels B,F), and the interior portion of the extended P (P1-DS04361 FISH signals—rhodamine channel, panels C,G). A close-up view of the pseudocolored overlay image (panel I) reveals the cytogenetic fine structure of the extended P element. The FISH images show good correspondence between the FISH staining patterns (I) and the structure of the extended P element (H; see Fig. 1).
Analysis of the cytogenetic substructure of the transposon at higher resolution (Fig. 3, panels E–H) revealed two more important features of ExtP1-7. First, differential retention of vector sequences in the corresponding half-P elements produced asymmetric labeling at the ends of the extended P element, allowing us to determine its orientation relative to the chromosome as 5HP distal, 3HP proximal. Second, after 3D image deconvolution, we consistently observed a gap in the FISH signals obtained with the P1 genomic probe and the 5′ end of ExtP1-7 (Fig. 3F). This result suggests that the genomic DNA cloned in the P1 phage spans only a portion of the region between the extended P ends, and it is consistent with our genomic map of the extended P-element donor at its original site on the third chromosome (Fig. 1). When viewed as a three-color projection (Fig. 3I), the fine-scale cytogenetic staining patterns indicate that the extended P element has retained its overall organization after transposition.
Molecular Integrity of ExtP1-7 on the X Chromosome
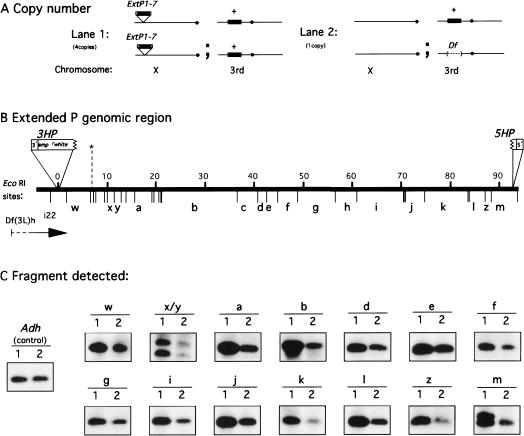
The structural integrity of the genomic DNA sequences contained within ExtP1-7 was examined at the molecular level by Southern blot analysis (Fig. 4). Using genomic DNA isolated from flies homozygous for the X-linked transposon and flies heterozygous for a deficiency spanning the region encompassed by the extended P element, we determined the relative genomic copy numbers for most of the EcoRI restriction fragments contained within the extended P element (Fig. 4A). Strains of the genotype shown in Fig. 4A (lane 1) are expected to harbor four copies of genomic sequences corresponding to the internal region of ExtP1. However, strains of the genotype shown in Fig. 4A (lane 2) are expected to harbor only one copy of genomic sequences corresponding to the internal region of ExtP1. Therefore, we predicted that if the extended P element were intact, we would see a genomic Southern hybridization signal ratio of 4 : 1 for any probe hybridizing to sequences within the extended P element. Hybridization with 12 independent DNA probes was quantified and produced approximately a 4 : 1 signal ratio for genomic EcoRI restriction fragments covering most of the genomic region spanned by the extended P element. The EcoRI fragments near the 3′ end (Fig. 4C, bands w,x/y,a,b) and near the 5′ end (Fig. 4C, bands j,k,l,z,m) had a copy number ratio (see Methods) average of 4.1 (range 3.5–4.3). However, the EcoRI fragments in the middle (Fig. 4C, bands d,e,f,g,i) had a copy number ratio average of 2.6 (range 2.1–3.5). The reduction in signal ratio observed corresponds to the genomic region containing chorion genes and may have resulted from differences in chorion gene amplification (Fig. 4B,C) (Ish-Horowicz et al. 1985; Delidakis and Kafatos 1987). To test this hypothesis, we also determined the copy number of this middle genomic region using hemizygous males in which chorion gene amplification does not occur; the expected copy number ratio should be 3:1. Consistent with this prediction, we found the copy number ratio for Figure 4, bands d,e,f,g, averaged 3.1 (range 2.9–3.2).
Figure 4.
Assessment of structural integrity of the X-linked extended P element ExtP1-7 by quantitative Southern analysis. (A) Genotypes of fly strains used for genomic copy-number comparison, shown with respect to the copy number of the genomic region encompassed by the extended P element (shown as a solid black bar) on the X chromosome (for ExtP1-7) and on the third chromosome. (B) The EcoRI restriction map of the genomic region encompassed by the extended P element. Genomic DNA is shown as a solid line with size kb above and the EcoRI restriction sites below. Specific EcoRI restriction fragments used for the copy-number comparison are labeled with bold letters a–m (Spradling 1981) and w–z. (C) Representative quantitative Southern hybridization results are shown for 16 EcoRI restriction fragments that were identified with 12 independent probes.
Deletions and internal rearrangements within ExtP1-7 would likely create detectable RFLPs. However, the conservation of restriction fragment sizes indicates that the transposition of the extended P did not result in gross sequence rearrangements within the transposed genomic region. Taken together, our molecular and cytogenetic analyses provide strong evidence that the extended P-element system is capable of producing large, stable transgenomic duplications.
Functional Integrity of ExtP1-7 on the X Chromosome
We next examined the conservation of function for both the transposable element sequences and the internal genomic DNA of ExtP1-7. We conducted crosses to determine whether ExtP1-7 could rescue, in trans, the lethal mutation l(3)rK561, which maps within the genomic region contained within the extended P element. If so, the wild-type gene within ExtP1-7 should rescue the chromosome 3 lethality. For testing genetic rescue of l(3)rK561 by the X-linked extended P element ExtP1-7, we crossed w1118ExtP1-7; Df(3L)hi22Ki pp/TM6B, Tb e males to l(3)rK561 ry/TM3 Sb ppe females. This cross produced 53 w1118ExtP1-7/+; Df(3L)hi22Ki pp/l(3)rK561 ry females but no +; Df(3L)hi22Ki pp/l(3)rK561 ry males. Because the lethality of the Df(3L)hi22/l(3) rK561 transheterozygotes is specifically rescued in the female progeny but not in the male progeny, we concluded that the ExtP1-7 retained genetic function for internal sequences. In other crosses, we could directly demonstrate cosegregation of the white marker associated with the X-linked ExtP1-7 and the rescuing activity of the X chromosome (data not shown).
Finally, we wanted to determine whether ExtP1-7 was itself capable of transposition. We therefore carried out a second transposition screen using ExtP1-7 as the donor. Transposition of ExtP1-7 was carried out by first crossing w1118ExtP1-7 homozygous females to w1118; P{ry+,Δ23}99B, Sb males. The resulting mosaic male progeny (w1118ExtP1-7; P{ry+, Δ23}99B, Sb/+) were then outcrossed to w1118 females. Because the extended P element is X linked, male progeny all inherit the w1118 chromosome, so autosomal transposition events can be identified as orange-eyed males. An autosomal transposition event was identified from among ∼2 × 104 male progeny (an estimated rate of 0.5 × 10−4), demonstrating that the extended P element, ExtP1-7, is still a functional transposon.
DISCUSSION
Our results provide proof of principle for the construction and mobilization of functional extended P elements and demonstrate that a 100-kb P element is capable of transposition to new sites in the Drosophila genome. The in vivo manipulation of half-transposons to produce large transposable elements provides an unprecedented approach for manipulating large eukaryotic genomic DNA fragments and could potentially be developed in other organisms containing appropriate transposable element vectors.
The method reported here is, to our knowledge, the first to be capable of moving large, defined chromosomal regions to unlinked sites in the Drosophila genome. Transposition of a naturally occurring TE element (Ising and Block 1984), composed of a large genomic region flanked by composite FB-NOF transposable elements (Harden and Ashburner 1990) has been investigated previously but is limited to the 3C2–6 region of the X chromosome and is not amenable to further manipulation. Autosynaptic methods can be used to create synthetic chromosomal deletions and duplications, but the resulting duplications are intrachromosomal (Gubb et al. 1988). Similarly, while site-specific recombination systems such as the FLP/FRT system have proven useful for genome manipulation, only small DNA fragments have been successfully transposed to unlinked insertion sites in the Drosophila genome (Golic et al. 1997). In contrast, we have demonstrated that large, defined chromosomal regions can be translocated to unlinked sites in the genome with the extended P elements.
Transposition Rates of ExtP1
We found that interchromosomal transposition of the ExtP1 occurs at a rate of approximately 1 in 10,000 flies. This value is consistent with the idea that transposition rates continue to decrease with increasing transposon size. Other factors can influence transposition rate, however.
First, because our extended P element is composed of half-P elements from different full-length P elements, the eight base-pair target-site duplications will not match, and this may have a deleterious effect on transposition rate. Second, because the half-P elements are produced as deletion derivatives of full-length P elements, the P-element sequences that are retained will be variable and may interfere with transposition. Last, we expect transposition rates for large P elements to be variable because P-element transposition appears to require interaction between the two P-element termini (Beall and Rio 1997). The frequency of such interactions is likely to be strongly influenced by the proximity of the two half-P-element ends within the nucleus and the higher-order chromatin structure of the genomic region containing the extended P element. However, we note that even with no further increases in transposition rate, a single researcher could easily identify a number of putative transposition events in a single day; thus, this method has the potential to be scaled up and applied to a larger proportion of the Drosophila genome. Further, the relatively larger size of extended P elements means that fewer such transposition events would be necessary for genome coverage. For example, at an average size of 100 kb, approximately 500 extended P elements would be required to collectively span each of the major autosomes, while at an average size of 500 kb, only about 100 extended P elements would be required.
We also note that the isolation and identification of half-P elements from full-length P elements is labor intensive and creates extended P elements whose ends may not be completely compatible for transposition. As a more efficient and reliable alternative approach, we are currently introducing specific, compatible 5′ and 3′ half-P elements into the Drosophila genome using germ-line transformation mediated by the piggyBac transposon vector, previously shown to be capable of high-frequency germ-line transformation and transposition in D. melanogaster (Handler and Harrell 1999). These half-P elements can then be mobilized to hundreds of genomic sites, and both the insertion site and orientation of each can be determined, comparing the genomic DNA sequences flanking each insertion with the available Drosophila genomic sequences. Appropriately oriented and compatible half-P elements can then be put together by simple meiotic recombination to produce a set of extended P elements that will allow us more rigorously to evaluate the relationship between P element size and transposition frequency.
Potential for Functional Genomics
Transpositional Duplications
Extended P elements can contribute to functional analysis of the Drosophila genome in a number of ways. We have demonstrated that extended P elements can be used to create interchromosomal transpositional duplications. The availability of such defined chromosomal duplications would allow rapid genetic mapping of mutations by complementation or transgene rescue and permit the identification of mutations in genes too large to be transformed by conventional methods.
An analogous approach, the introduction of large fragments by transformation, has been proposed in a number of eukaryotes, including Drosophila. Most recently, this approach has been proposed in the zebrafish, D. rerio, using the transposable element Tc3 to introduce large genomic fragments that could then be used to map the growing set of chemically induced mutations by complementation (Izsvak et al. 1997). Our method provides an alternative in vivo approach for the production of large transgenomic regions for genetic mapping by complementation.
Excisions and Internal Deletions
It is well known that imprecise excision of P elements creates internally deleted P elements at very high frequencies (Engels 1996). Many such excisions leave behind little more than the extreme P element termini, while others retain variable amounts of the P internal sequences. Of the 35 ExtP1 excisions we characterized, we have found that 31 (88.6%) fail to complement the l(3)rK561 mutation near the center of the genomic region encompassed by the transposon, demonstrating the loss of genomic sequences between the P ends. By analogy to chromosomal deletions created by excision of two full-length P elements in the cis-configuration (Cooley et al. 1990), we expect that most of these excision events have lost all the genomic sequences between the P-element ends, the very same genomic sequences that underwent transposition to new sites in the genome. Conversely, the observation that four of the 35 excision events complement the lethal mutation is consistent with the production of internal deletions that occur as a result of the excision/repair process and retain part of the genomic region between the P-element ends. This raises the possibility of carrying out direct mapping of lethal complementation groups within extended P elements by genetic and molecular analysis of internal deletions retaining different complementation groups.
Matching Deletions and Duplications
The simultaneous use of complementary duplications and deletions can be used to provide a reliable, accurate, and efficient approach for genetic mapping. Specifically, mutations mapping within an extended P element would be rescued by the transpositional duplication of the genomic region contained within the extended P element and would fail to be complemented by the matching deletion derivative of the same region, potentially in a single genetic cross. Such fly strains, containing large deletions and matching transpositional duplications, would be stable as long as no genetic functions are disrupted at the breakpoints or insertion site. The production of a set of matching duplications and deletions covering a significant proportion of the Drosophila genome would represent a new type of in vivo large-fragment library in which individual genomic fragments could be added or removed by conventional genetic crosses. Using such a library, large sets of mutations could be rapidly mapped in a single cross to specific genomic regions defined by transpositional duplications and deletions created using extended P elements.
Potential for Investigation of Genetic and Epigenetic Phenomena
Extended P elements also create new avenues for experimental analysis of chromatin structure, gene regulation, and position effect. For example, the observed reductions in chorion gene amplification within ExtP1-7 suggested by our analysis of genomic copy number are consistent with earlier results with relatively small P-element transgenes that indicated the existence of long-range negative position effects (Delidakis and Kafatos 1987). Our results extend these previous ones by showing that negative position effects can be obtained even when the entire amplified region is included and suggest that either chromatin boundaries are outside of this large region or that other as yet undefined mechanisms operate to insulate the chorion genes from long-range negative position effects at their normal location on the third chromosome.
Evolutionary Implications of Extended P Elements
Last, we note that P transposase activity produces half-P elements at appreciable frequencies (Svoboda et al. 1995), making it likely that many such half-P elements exist in natural populations of Drosophila. The demonstration that extended P elements can be created by in vivo manipulation in the laboratory and that the resulting transposons are capable of undergoing excision and transposition further suggests that extended P elements may also be produced in natural populations. While the relatively recent invasion of P elements into D. melanogaster (Engels 1996) makes it unlikely that extended P elements or their derivatives could be easily identified in natural populations, our results are consistent with the idea that extended P elements are likely to play a role in Drosophila genome evolution. In addition, our results suggest that half-transposons previously considered to be inert remnants of functional transposable elements may be used for the creation of new functional transposons with the potential to alter the structure of the genome.
METHODS
Nomenclature of P Elements and Derivatives Used in This Study
In describing the in vivo construction of the extended P element, the progenitor P elements (described below) are referred to as donor P elements and the resulting deletion derivatives that have lost the function of one end are referred to as half-P elements. The intact P element used to generate the 5′ half-P element is referred to as the 5DP, for 5′ donor-P element, and the resulting deletion derivative is referred to as the 5HP, for 5′ half-P element because it retains a functional 5′ end. Likewise, the progenitor and deletion derivative of the P element used to generate the 3′ half-P element are referred to as the 3DP (for 3′ donor-P element) and the 3HP (for 3′ half-P element), respectively. The extended P element (described below) on chromosome 3L is referred to as ExtP1. Transposition events are numbered sequentially as ExtP1-1, ExtP1-2, and so forth. The ExtP1-7, examined in detail in this report, is the transposed extended P element located on the X chromosome (below).
Construction of the 3HP on Chromosome 3L
Strains containing single P{lacW} transposon insertions (Bier et al. 1990) were generous gifts from the laboratory of M.P. Scott (Stanford University). Individual strains were crossed to a strain carrying a stable genomic source of P transposase, P{ry+, Δ23}99B, that is active in both germ-line and somatic tissues (Robertson et al. 1988). Single mosaic F1 flies were then crossed back to flies bearing the P{ry, Δ23}99B transposase source, and the F2 flies screened for orange-eyed individuals that are no longer mosaic in the presence of P{ry, Δ23}99B, indicating that they contain a functional white gene but can no longer respond to P transposase. Twenty-eight nonmosaic strains were retained, and the presence of P-element ends assessed by Southern blot hybridization with P-element end-specific probes. The probes were produced by PCR using a primer common to both ends (called PRI, 5′CAT GATGAAATAACATAAGGTGGTCCCGTCG3′) in combination with either a 5′ end-specific primer, 5′GGCTGCACCCAAGGCTCTGCTCCCACA3′ or a 3′ end-specific primer, 5′CCCCACGGACATGCTAAGGGTTAAT3′. These primer pairs were used to produce either a 260-bp 5′-end-specific probe or a 158-bp 3′-end-specific probe.
Identification of a Compatible 5DP on Chromosome 3L
Strains containing putative P-element insertions proximal to the 3HP element were obtained from available stock collections (BDGP, Indiana Stock Center) and by local transposition of the 3DP element (Tower et al. 1993; Zhang and Spradling 1993; Golic 1994). Strains obtained by local hopping were further characterized by determination of their meiotic map distance from the 3HP element. Those giving recombination rates of <1% were retained for further analysis. Candidate strains were next characterized by in situ hybridization to polytene chromosomes and hybridization of genomic DNA flanking the insertion sites to an ordered array of P1 genomic clones (constructed by our laboratory on the basis of in situ hybridization data available from the BDGP).
To identify and orient P-element insertions contained within the Cp15 genomic DNA contig (BDGP), we further characterized strains containing P-element insertions mapping to the 66DE region by hybridization of restriction-digested P1 clone DNA with genomic DNA probes flanking the P-element insertions, leading to the identification of the 5DP element P{lacW}-j5B6. The insertion site and orientation of the 5DP element was confirmed by the identification of a previously defined sequence-tagged site, sts631 (BDGP, see Fig. 1) beginning 101 bp away from its 3′ end.
Production and Confirmation of the Extended P Element, ExtP1
The low frequency with which half-P elements retaining the white gene were obtained in the genetic screen for loss of somatic mosaicism, as well as the identification of other P-element fusion derivatives containing no 5′ ends, but multiple 3′ ends (data not shown) suggested that half-P derivatives are produced by multistep processes. As asymmetric repair is likely to be distance dependent, we hypothesized that smaller half-P-element deletion derivatives that had lost the white gene could be obtained at higher frequencies. We therefore decided to produce 5′ half-P-element partners for the 3HP element using a screening method that would allow identification of the desired extended P element without requiring that the resulting 5HP retain a functional white gene (Fig. 2).
The 5DP element (P{lacW}-j5B6) and 3HP (P159.1) were first recombined into a cis configuration by meiotic recombination and the recombinant chromosome confirmed by Southern blot analysis (Fig. 2, lane 3). This strain was then crossed to the P-transposase-producing strain. Eye-color mosaicism was seen in the F1 progeny because of transposition and excision of the 5DP element (not shown). These mosaic progeny were then outcrossed to a w1118 strain, and excision events were identified by screening for changes in eye color from red-eyed flies (two copies of the white gene) to orange-eyed flies (one copy of the white gene). Southern blot analysis of 135 independent excision/deletion (orange-eyed) derivative strains was carried out with the P-element end-specific probes (see above). This led to the identification of 22 P-element deletion derivatives that had retained the 5′ end and lost the 3′ end of the 5DP element while maintaining the original structure of the 3HP element. One such strain contained the 5HP derivative, which appeared to have lost the EcoRI sites present within the 5DP element. Hybridization with genomic DNA probes flanking the insertion indicated that approximately 1 kb of the P element remained at the insertion site. PCR amplification of the 5HP element using primers that would anneal to genomic sequences flanking the original 5DP insertion site was performed to further examine the structure of the 5HP derivative. The sequence of the distal primer (3′ end of the 5DP element) was 5′ATCACTAATCCGCCACGTTGG3′, and the sequence of the proximal primer (5′ end of the 5DP element) was 5′CTGAGCGAAGTTTGGTTTTTGATTCC3′. The resulting 1.3-kb PCR product was cloned into the pPCR2.1 TA cloning kit (Invitrogen) and sequenced.
Size Determination of ExtP1
The initial size estimate of the entire ExtP1 on chromosome 3L was based on the overlapping EcoRI restriction maps available for the two genomic clone walks described (Spradling 1981; Ish-Horowicz et al. 1985) and agrees with more recent and more accurate estimates based on comparisons with available genomic sequence (BDGP and Celera, CSC:AC014798). The available ordered genomic DNA sequence for this genomic region begins approximately 6.5 kb proximal to the 3HP insertion site, whereas the 5HP element is located at coordinate 84,500 on the genomic sequence contig. This predicts a size for this genomic region of ∼91.0 kb and an overall size of ∼100 kb, including the P-element vector sequences at both ends, as shown in Figure 4.
Analysis of Occurrence and Rates of Transposition of ExtP1
The ExtP1 strain was crossed to the P-transposase-producing strain, and the resulting orange-eyed mosaic males (ExtP1/P{ry+, Δ23}99B, Sb) were mated to w1118; TM3/TM6B females (Lindsley and Zimm 1992). Eighty red-eyed progeny were identified among ∼4 × 104 progeny. Two putative interchromosomal transposition events were identified by crossing each red-eyed fly individually to a w1118; ′TM3, Sb e/TM6B, Tb Hu e strain. Segregation of orange eye color in the following generation was used as an indicator of the independent assortment of two white genes. Because only one-half the progeny in the original screen for red-eyed progeny receive the donor ExtP1-bearing chromosome, we estimate the interchromosomal transposition rate as 2/(0.5 × 4) × 10−4 =1 × 10−4, or 0.01%. We also note that a higher proportion of the putative transpositions appear to be intrachromosomal than normally observed, suggesting that there may be increases in the relative rates of local transposition (Tower et al. 1993; Zhang and Spradling 1993; Golic 1994) and localized rearrangements (Delattre et al. 1995) with increasing P-element size.
Cytogenetic Analysis of ExtP1-7 on the X Chromosome
Salivary gland polytene chromosome squashes were prepared for in situ hybridization as previously described (Spradling 1981). FISH probes (of EcoRI-linearized plasmid pP{LacW} or P1 [DS04361]) were random-primed labeled with the fluorescent nucleotides Alexa-488-dUTP (Molecular Probes) or FluoroRed dUTP (Amersham) according to the procedure described by Bass et al. (2000). Labeled DNAs were purified and redissolved in hybridization buffer (50% formamide, 5× SSC, 5× Denhardt's, 0.1% SDS) at a concentration of 15 μg/mL of P1 probe and 0.7 μg/mL of pP{LacW}. Chromosome squashes were incubated with 30 μL of probe at 37°C for 16 h, stained with DAPI, and mounted in VectaShield (Vector labs). All images were recorded with an Olympus IMT-2 wide-field deconvolution microscope workstation (DeltaVision, Applied Precision) with an oil-immersion lens (60× NA 1.4 PlanApo, Olympus). Optical reconstructions (0.3 μ Z-step) were subjected to three-dimensional iterative deconvolution (Chen et al. 1995) and displayed as projections spanning four to six optical sections each (Fig. 3).
Molecular and Copy Number Analysis of ExtP1-7 on the X Chromosome
DNA for quantitative Southern blot analysis was isolated from females of the genotype w1118 ExtP1-7/w1118 ExtP1-7 and females of the genotype Df(3L)hi22Ki pp/TM3, Sb e. We used females exclusively in order to maximize the copy number of the extended P element on the X chromosome (Spradling 1981). The deficiency Df(3L)hi22 is expected to delete all genomic sequences contained within the extended P element (Ish-Horowicz et al. 1985). DNA probes used for quantitative Southern blot analysis were derived from a number of sources. For the hairy genomic region, we used genomic fragments obtained by plasmid rescue (Ashburner 1989) of the 3DP and 3HP elements and a cosmid, cosh spanning most of the hairy genomic region (Ish-Horowicz et al. 1985; Rushlow et al. 1989). The proximal genomic region contains EcoRI genomic restriction fragments a through m (Spradling 1981). Genomic DNA for probes corresponding to each fragment were as follows: Fragments a and b were isolated from genomic λ phage generously provided by the laboratory of A. Spradling. Fragments c, d, e, and f were obtained by plasmid rescue of genomic DNA flanking the l(3)rK561 P-element insertion. Fragments g through m were obtained by PCR amplification of specific genomic regions spanning the junctions between adjacent EcoRI restriction fragments. Specific genomic DNA sequences used for the selection of primers were based on the available genomic sequence for the region (CSC:AC014798, BDGP, and Celera). Sequences of primers used for copy number analysis are available on request. Primer pairs were chosen so that the amplified fragment-based probes would span two or more specific EcoRI sites. In this way, PCR products could be used simultaneously to assess the copy number and integrity of two or more EcoRI restriction fragments. Hybridization signals were quantified by means of a phosphoimager (Molecular Dynamics) using the ImageQuant 4.2 software (Molecular Dynamics). Multiple exposures were used to determine the linear dose-response range for each blot. Hybridization signal values from linear-range exposures were subsequently normalized to a second single-copy reference signal (Adh1 gene, GenBank DMADHGC) obtained by stripping and reprobing of blots. The relative copy number obtained (see Results) for each fragment was determined according to the formula (lane 1probe/lane 2probe)/(lane 1Adh/lane 2Adh).
Acknowledgments
We thank M. Scott and members of the Scott lab, Kathy Matthews and the Indiana Stock Center, and the Berkeley Drosophila Genome Project for providing fly stocks; A. Spradling (Carnegie Institute of Washington) and D. Ish-Horowicz (Imperial Cancer Research Fund) for providing genomic clones in the 66D/E region; Chris Bacot and the FSU DNA sequencing facility for DNA sequencing; Cathy Trivigno, Sameer Khatri, and Wesley Dobbs for their assistance in identifying local P-element transposition events and P-element excision events; Steve Stowers for suggesting the use of quantitative Southerns; M. Hurt for helpful comments on the manuscript; A. Thistle for editorial help; and K. Womble for technical assistance with figures. This work was supported by the National Institutes of Health (D.G., NIH-HG01583) and the Florida State University Research Foundation—Program Enhancement Grant (H.W.B.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL garza@bio.fsu.edu; FAX (850) 644-0481.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.151700.
REFERENCES
- Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Bass HW, Riera-Lizarazu O, Ananiev EV, Bordoli SJ, Rines HW, Phillips RL, Sedat JW, Agard DA, Cande WZ. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J Cell Sci. 2000;113:1033–1042. doi: 10.1242/jcs.113.6.1033. [DOI] [PubMed] [Google Scholar]
- Beall EL, Rio DC. Drosophila IRBP/Ku p70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes & Dev. 1996;10:921–933. doi: 10.1101/gad.10.8.921. [DOI] [PubMed] [Google Scholar]
- ———. Drosophila P-element transposase is a novel site-specific endonuclease. Genes & Dev. 11: 2137–2151. [DOI] [PMC free article] [PubMed]
- Bier E, Jan LY, Jan YN. rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes & Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- Bilder D, Scott MP. Genomic regions required for morphogenesis of the Drosophila embryonic midgut. Genetics. 1995;141:1087–1100. doi: 10.1093/genetics/141.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Swedlow JR, Grote M, Sedat JW, Agard DA. The collection, processing, and display of digital three-dimensional images of biological specimens. In: Pawley JB, editor. Handbook of biological confocal microscopy. New York: Plenum; 1995. pp. 197–210. [Google Scholar]
- Cooley L, Thompson D, Spradling A C. Constructing deletions with defined endpoints in Drosophila. Proc Natl Acad Sci. 1990;87:3170–3173. doi: 10.1073/pnas.87.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Anxolabehere D, Coen D. Prevalence of localized rearrangements vs. transpositions among events induced by Drosophila P element transposase on a P transgene. Genetics. 1995;141:1407–1424. doi: 10.1093/genetics/141.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C, Kafatos F. Amplification of a chorion gene cluster in Drosophila is subject to multiple cis-regulatory elements and to long-range position effects. J Mol Biol. 1987;197:11–26. doi: 10.1016/0022-2836(87)90605-x. [DOI] [PubMed] [Google Scholar]
- Engels WR. P elements in Drosophila. In: Saedler H, Gierl A, editors. Transposable elements. Berlin: Springer; 1996. pp. 103–123. [Google Scholar]
- Golic KG. Local transposition of P elements in Drosophila melanogaster and recombination between duplicated elements using a site-specific recombinase. Genetics. 1994;137:551–563. doi: 10.1093/genetics/137.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic MM, Rong YS, Petersen RB, Lindquist SL, Golic KG. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 1997;15:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubb D, McGill S, Ashburner M. A selective screen to recover chromosomal deletions and duplications in Drosophila melanogaster. Genetics. 1988;119:377–390. doi: 10.1093/genetics/119.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenlin M, Steller H, Pirrotta V, Mohier E. A 43 kilobase cosmid P transposon rescues the fs(1)K10 morphogenetic locus and three adjacent Drosophila developmental mutants. Cell. 1985;40:827–837. doi: 10.1016/0092-8674(85)90342-3. [DOI] [PubMed] [Google Scholar]
- Handler AM, Harrell RA., II Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8:449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- Harden N, Ashburner M. Characterization of the FB-NOF transposable element of Drosophila melanogaster. Genetics. 1990;126:387–400. doi: 10.1093/genetics/126.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D, Howard KR, Pinchin SM, Ingham P. Molecular and genetic analysis of the hairy locus in Drosophila. Cold Spring Harbor Symp Quant Biol. 1985;50:135–144. doi: 10.1101/sqb.1985.050.01.019. [DOI] [PubMed] [Google Scholar]
- Ising G, Block K. A transposon as a cytogenetic marker in Drosophila melanogaster. Mol Gen Genet. 1984;196:6–16. doi: 10.1007/BF00334085. [DOI] [PubMed] [Google Scholar]
- Izsvak Z, Ivics Z, Hackett PB. Repetitive elements and their genetic applications in zebrafish. Biochem Cell Biol. 1997;75:507–523. [PubMed] [Google Scholar]
- Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. San Diego: Academic Press; 1992. [Google Scholar]
- Mullins MC, Rio D, Rubin GM. cis-acting DNA sequence requirements for P-element transposition. Genes & Dev. 1989;3:729–738. doi: 10.1101/gad.3.5.729. [DOI] [PubMed] [Google Scholar]
- O'Connor M, Peifer M, Bender W. Construction of large DNA segments in Escherichia coli. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- O'Hare K, Rubin GM. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983;34:25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Rio DC, Rubin GM. Identification and purification of a Drosophila protein that binds to the terminal 31-base-pair inverted repeats of the P transposable element. Proc Natl Acad Sci. 1988;85:8929–8933. doi: 10.1073/pnas.85.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow CA, Hogan A, Pinchin SM, Howe KM, Lardelli M, Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 1989;8:3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, King JA, Orr-Weaver TL. Identification of genomic regions required for DNA replication during Drosophila embryogenesis. Genetics. 1993;135:817–829. doi: 10.1093/genetics/135.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. The organization and amplification of two chromosomal domains containing Drosophila chorion genes. Cell. 1981;27:193–201. doi: 10.1016/0092-8674(81)90373-1. [DOI] [PubMed] [Google Scholar]
- ————— . P element mediated transformation. In: Roberts DB, editor. Drosophila: A practical approach. Oxford: IRL; 1986. pp. 175–197. [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehling-Hampton K, Ciampa PJ, Brook A, Dyson N. A genetic screen for modifiers of E2F in Drosophila melanogaster. Genetics. 1999;153:275–287. doi: 10.1093/genetics/153.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda YHM, Robson MK, Sved JA. P-element-induced male recombination can be produced in Drosophila melanogaster by combining end-deficient elements in trans. Genetics. 1995;139:1601–1610. doi: 10.1093/genetics/139.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z, Bingham PM. Regulation of white locus expression: The structure of mutant alleles at the white locus of Drosophila melanogaster. Cell. 1982;30:529–541. doi: 10.1016/0092-8674(82)90250-1. [DOI] [PubMed] [Google Scholar]
- Zhang P, Spradling AC. Efficient and dispersed local P element transposition from Drosophila females. Genetics. 1993;133:361–373. doi: 10.1093/genetics/133.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]