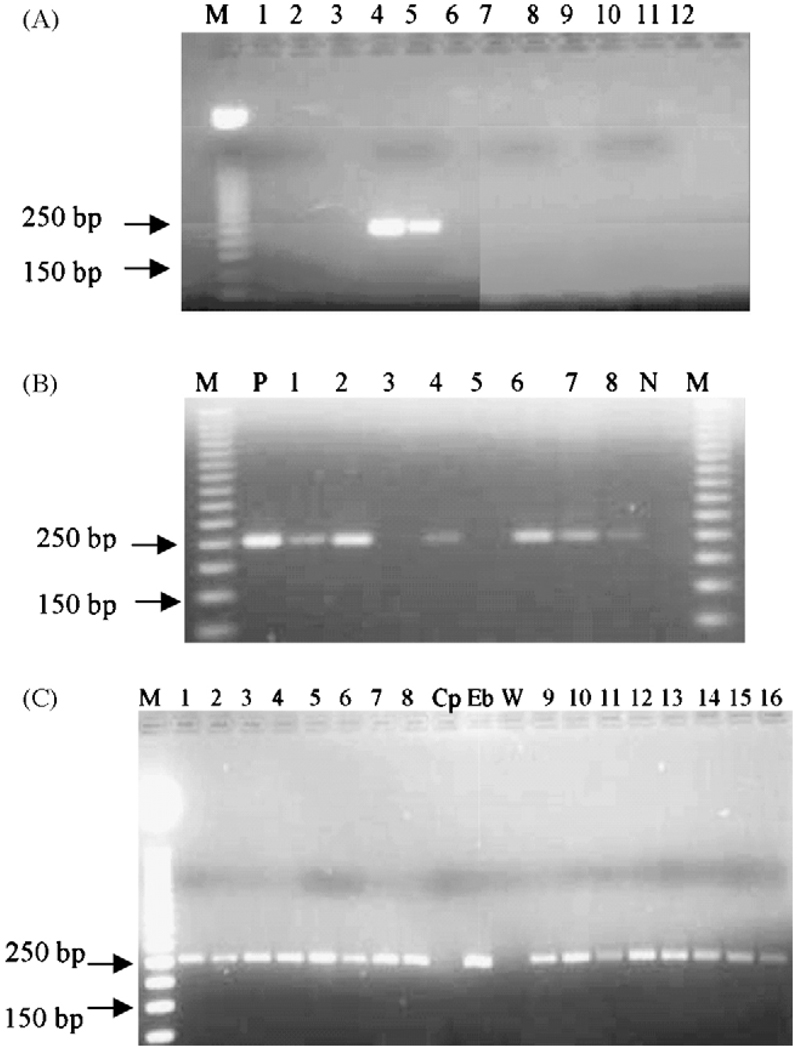
Figure 1.
(A) Confirmation of the specificity of the primers: the primers were tested for specificity in a PCR reaction using water (lane 1), DNA extracted from Cryptosporidium parvum (lane 2) and C. hominis oocysts (lane 3), Entamoeba histolytica DNA (lane 7), Ent. dispar (lane 8), Campylobacter DNA (lane 10), human DNA (lane 11) and two different concentrations of Enterocytozoon bieneusi DNA (lane 4: 3 ng/ml; lane 5: 0.3 ng/ml). Lanes 6, 9 and 12 were left empty. M: molecular marker (50 bp ladder; Promega, Madison, WI, USA). (B) Agarose gel of PCR reaction using genomic DNA extracted from stool samples, positive control (DNA from E. bieneusi oocysts: lane P), negative control (Cryptosporidium DNA: lane N) followed by restriction digest with PstI. Lanes 3 and 5 were two negative stools and lanes 1, 2, 4, 6, 7 and 8 were positive samples. There was no change (no digestion) indicating that the organism was E. bieneusi as recommended by Fedorko et al. (1995). (C) Comparison of PCR products from DNA purified from stool samples positive by microscopy, using the alkaline digestion with KOH and dithiotheithrol (lanes 1 to 8) and the same samples treated by glass bead disruption (lanes 9 to 16). Cryptosporidium parvum DNA (Cp) and water (W) were used as negative controls; E. bieneusi (Eb) DNA was used as the positive control.