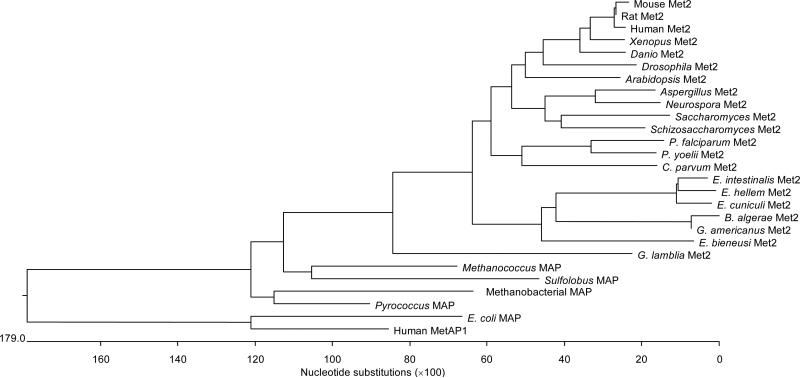
Fig. 3.
Clustal_W analysis of MetAP2, MetAP1 and MAP proteins using MegAlign (DNASTAR). MetAP2: Xenopus Met2 (Xenopus laevis), Schizosaccharomyces Met2 (Schizosaccharomyces pombe), Saccharomyces Met2 (Saccharomyces cerevisiae), Rat Met2 (Rattus norvegicus), P. yoelii Met2 (Plasmodium yoelii), P. falciparum Met2 (Plasmodium falciparum), Neurospora Met2 (Neurospora crassa), Mouse Met2 (Mus musculus), Human Met2 (Homo sapiens), G. lamblia Met2 (Giardia lamblia), Drosophila Met2 (Drosophila melanogaster), Danio Met2 (Danio rerio), C. parvum Met2 (Cryptosporidium parvum), Aspergillus Met2 (Aspergillus nidulans), Arabidopsis Met2 (Arabidopsis thaliana), B. algerae Met2 (Brachiola algerae), G. americanus Met2 (Glugea americanus), E. cuniculi Met2 (Encephalitozoon cuniculi), E. intestinalis Met2 (Encephalitozoon intestinalis), E. hellem Met2 (Encephalitozoon hellem), and E. bieneusi Met2 (Enterocytozoon bieneusi). MetAP1: Homo sapiens. MAP (Peptidase M) Eubacteria: E. coli MAP (Escherichia coli). MAP (Peptidase M) Archaea: Sulfolobus MAP (Sulfolobus solfataricus P2), Pyrococcus MAP (Pyrococcus horikoshii), Methanococcus MAP (Methanococcus maripaludis S2), Methanobacterial MAP (Methanothermobacter thermautotrophicus).