Figure 2.
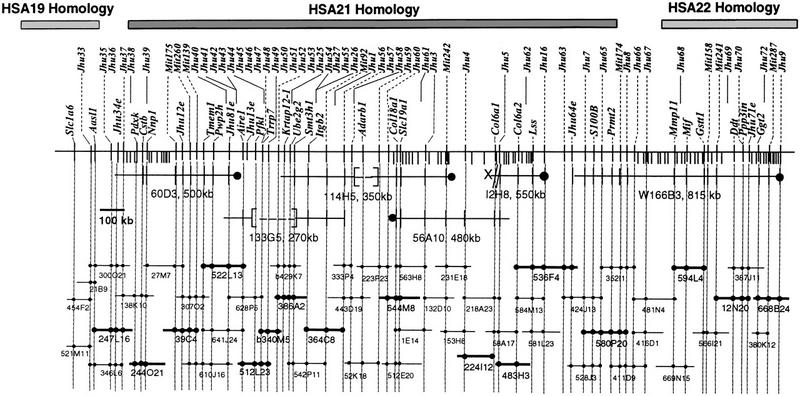
PAC-based STS content map of the region of MMU10 that shows conserved linkage with HSA19, HSA21, and HSA22. The contig spans 2.8 Mb and includes the entire HSA21 homologous region. Fifty-two of 156 clones are shown with addresses in the RPCI-21 or RPCI-22 libraries (BACs from the RPCI-22 library have the prefix ,“b”). STS markers are shown above and transcripts adjacent to the chromosome. The prefix “D10” has been omitted for Jhu and Mit STS markers. Six YACs that were used as hybridization probes are shown: [—-] indicates a deleted region, X-// a chimeric end, and filled circles the YAC centromeric ends. YAC fragmentation events are indicated as short lines extending beneath the main marker line (Cole et al. 1999). Clones are drawn to scale. Presence of a marker is indicated with a filled circle (a subset of the STS markers are shown here). Clones of the minimal tiling path that are currently being sequenced are indicated by bold lines. Sequence is available for clones 340M5 (GenBank accession no. AC007433), 644M8 (accession no. AC007641), 536F4 (accession no. AC007937), and 580P20 (accession no. AC006507).
