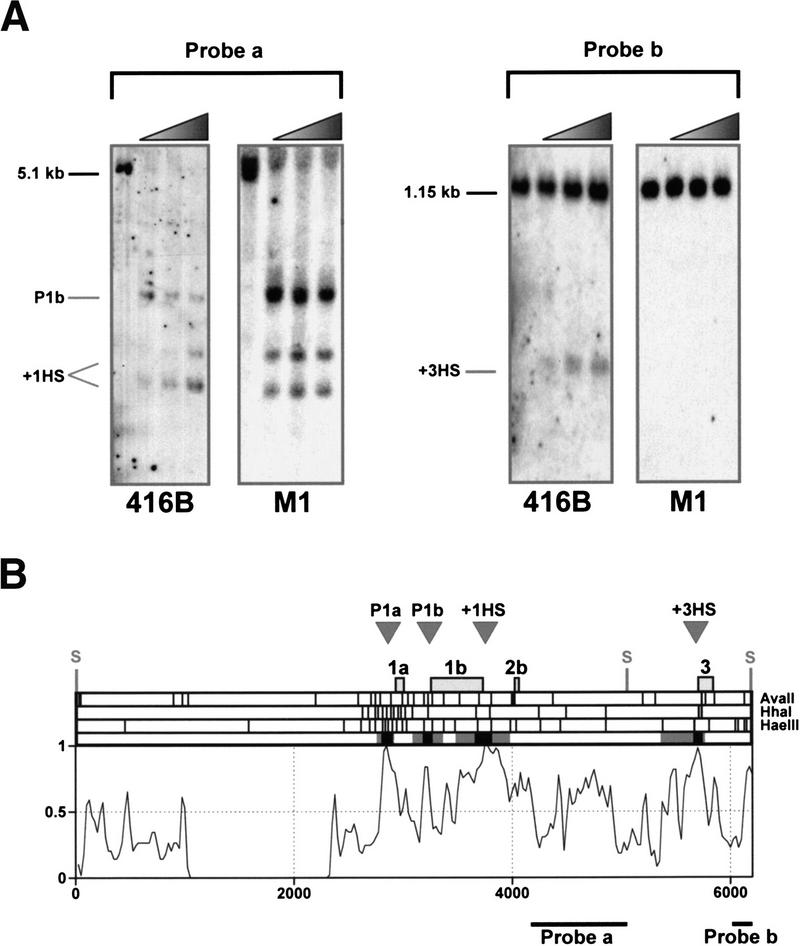
Figure 4.
Localized regions of sensitivity to restriction endonucleases correspond to peaks of human/mouse homology. (A) Restriction endonuclease accessibility assay showing the mapping of the hypersensitive sites at promoter 1b, +1 (+1HS), and +3 (+3HS) (numbering corresponds to distances in kb from the start of exon 1a) in the 416B and M1 primitive myeloid cell lines. Nuclei were incubated with HaeIII. DNA was subsequently extracted, digested with SacI, and hybridized with probes indicated in B. The absence of the +3 hypersensitive site in M1 is consistent with previous DNaseI hypersensitive site analysis (Göttgens et al. 1997). (B) Summary of restriction endonuclease data for the 5′ region of the mouse SCL gene. The top part of the diagram shows the approximate locations of previously mapped DNaseI hypersensitive sites (gray arrowheads labeled P1a, P1b, +1HS, and +3HS) followed by the positions of mouse SCL exons 1a to 3 and the SacI sites used for the Southern blots shown in part A. This is followed by restriction maps for the three enzymes (AvaII, HhaI, and HaeIII) used to determine endonuclease sensitivity, and a summary of the endonuclease sensitivity experiments in which black and gray boxes represent the minimum and maximum regions of endonuclease accessibility in 416B and/or M1 cells. The profile of the mouse/human alignment underneath is a 6250 nucleotide section of the alignment from Fig. 3 (see yellow bar in Fig. 3) and shows the concordance of endonuclease accessibility and sequence conservation.