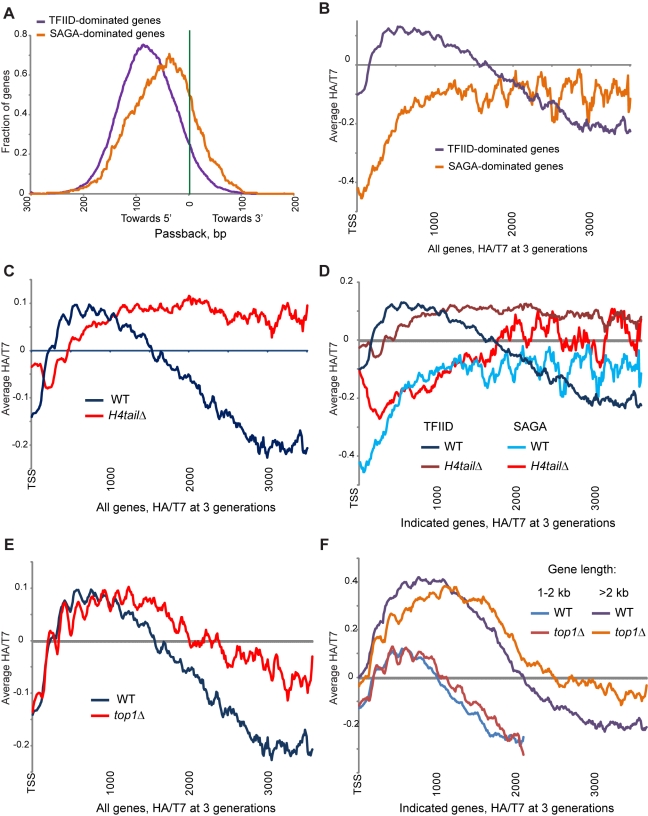
Figure 7. Mutants affecting ancestral histone retention.
(A) Distribution of lateral nucleosome distances from model (Figure 5). Shown are the passback parameters for SAGA-dominated and TFIID-dominated genes as defined in Huisinga et al. [38]. (B) TFIID-dominated genes preferentially accumulate 5′ H3-HA. Averages of 3 generation experimental data are shown for the indicated gene classes. (C) H4 tail deletion dramatically reshapes the landscape of ancestral histone retention. Yeast carrying an N-terminal H4 tail deletion were processed as in Figure 1A–B, and averages for all genes are plotted as indicated. We note that this strain has retained a wild-type HHT2-HHF2 locus for viability, so results must be interpreted with caution. However, we find similar but less dramatic effects in an H4K5,12R mutant (Figure S8B), supporting the observation here that passback is affected by the H4 N-terminal tail. (D) H4 tail deletion preferentially affects TFIID-dominated genes. Data for wild-type and H4tailΔ yeast are plotted for the indicated gene classes. (E) Topoisomerase I plays a role in 5′ accumulation of ancestral histones. top1Δ yeast were processed as in Figure 1A–B, and averages for all genes are plotted as indicated. (F) TOP1 deletion affects 5′ passback preferentially at long genes. Data for wild-type and top1Δ yeast are plotted for the indicated gene classes.