Abstract
The synthetic lethality screen is a powerful genetic method for unraveling functional interactions between proteins in yeast. Here we demonstrate the feasibility of a chemical synthetic lethality screen in cultured human cells, based in part on the concept of the yeast method. The technology employs both an immortalized human cell line, deficient in a gene of interest, which is complemented by an episomal survival plasmid expressing the gene of interest, and the use of a novel double-label fluorescence system. Selective pressure imposed by any one of several synthetic lethal metabolic inhibitors prevented the spontaneous loss of the episomal survival plasmid. Retention or loss over time of this plasmid could be sensitively detected in a blind test, while cells were grown in microtiter plates. Application of this method should thus permit high throughput screening of drugs, which are synthetically lethal with any mutant human gene of interest, whose normal counterpart can be expressed. This usage is particularly attractive for the search of drugs, which kill malignant cells in a gene-specific manner, based on their predetermined cellular genotype. Moreover, by replacing the chemicals used in this example with a library of either DNA oligonucleotides or expressible dominant negative genetic elements, one should be able to identify synthetic lethal human genes.
With continued progress in the Human Genome Project and the sequencing of the majority of the human cDNAs, use of functional genomic techniques for the elucidation of gene function has become a major priority (Lander 1996; Fields 1997). Previously, we have initiated the use of expressible human cDNA libraries for complementation (Teitz et al. 1987, 1989) or suppression (Teitz et al. 1990) of DNA damage-repair deficiency in inborn mutant human cells. In an effort to expand the arsenal of genetic tools that can be applied to human biological systems, we sought to introduce the concept of the yeast synthetic lethality screen (Bender and Pringle 1991) in human cells. The essence of this screen in yeast is the ability to identify nonallelic and nonessential mutations that are lethal in combination with a nonessential mutation in a gene of interest (i.e., synthetic lethality). A wild-type copy of the gene of interest, on an episomal plasmid, is introduced into cells that are null for expression of this gene. Random mutagenesis of the entire yeast genome within these cells may inactivate a gene that is synthetically lethal with the gene of interest. Under these conditions, retention of the episomal plasmid, which is otherwise spontaneously lost, and expression of the gene of interest becomes essential for survival (i.e., survival plasmid). Plasmid loss or retention is detected by changes in colony pigmentation. This genetic method is very powerful as it can reveal not only interactions between gene products with direct physical contacts but also interactions along the same or parallel pathways.
Inactivation of a specific gene product by a drug is in practical terms equivalent to its ablation by mutagenesis. We therefore sought to establish the principles of the method in human cells. This could be attained by the generation of synthetic lethal conditions via the use of drugs that inhibit particular enzymatic activity in lieu of genetic manipulation. Such a drug would then cause retention of an episomal survival plasmid encoding a gene activity that becomes essential for cell survival. An approach of this kind would allow screening of drugs that are synthetically lethal with a particular mutant gene, implying an ability to kill cells that carry such mutations.
To adapt this assay to human cells, we chose to use the Epstein-Barr virus (EBV)–based episomal vectors, which can replicate autonomously as a low copy number episome in human cells of diverse tissues (Yates et al. 1985). Retention of these vectors in human cells requires the continued selection for a dominant selectable marker gene built into the vector (Reisman et al. 1985; Calos 1996; N. Dafni and D. Canaani, unpubl.); otherwise, they are spontaneously lost over time. This spontaneous gradual plasmid loss can be taken advantage of by creating synthetic lethal conditions under which retention of an episomal plasmid expressing the gene of interest becomes indispensable for viability.
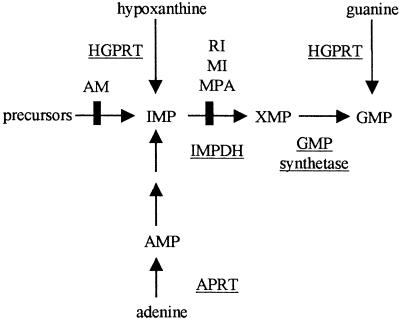
As a model system for the establishment of the method, the biosynthetic pathway leading to the production of guanosine monophosphate (GMP) was chosen (Fig. 1). This pathway is particularly amenable for a synthetic lethality screen as it involves multiple alternative paths to GMP biosynthesis. Under normal circumstances, GMP is synthesized through the de novo pathway, while the salvage pathways convert hypoxanthine and guanine to inosine monophosphate (IMP) and GMP, respectively, making hypoxanthine–guanine phosphoribosyl transferase (HPRT1) nonessential for cell survival. Thus, retention of an episome encoding HPRT1 cDNA, in an HPRT1-deficient human cell line, should become necessary for survival when the de novo biosynthetic pathway is blocked. To be able to follow loss or retention of an HPRT1 survival plasmid, we have used a novel double-label GFP fluorescent system. Several drugs, such as mycophenolic acid (MPA), which inhibits IMP dehydrogenase (IMPDH) enzyme activity within this metabolic pathway (Fig. 1), were used to create synthetic lethal conditions with concomitant retention of the survival plasmid (i.e., chemical synthetic lethality). Growth of cells in microtiter plates and use of a microplate fluorescence reader was utilized to follow fluorescence over time. This novel system should, in principle, allow high-throughput screening of reagents that are synthetically lethal with any mutant human gene of interest whose wild-type counterpart is expressible.
Figure 1.
De novo and salvage pathways of purine biosynthesis. Key enzymes are underlined. Arrows indicate the action of enzymes. The sites of inhibition by mycophenolic acid (MPA), ribavirin (RI), mizoribine (MI), and aminopterin (AM) are marked.
RESULTS
Design of the Human Synthetic Lethality System
To establish synthetic lethality as a high-throughput screening system based on fluorescent readout, it is essential to have a fluorescent marker that is expressed from the episomal survival plasmid. The strength of the fluorescent signal would then be correlated to the number of episomes present in the cell. However, this signal needs to be normalized relative to cell number. Ideally, the unstable presence of the episome and its marker could be measured against a second marker that is stably expressed in all cells. We chose the sapphire and topaz variants of the green fluorescent protein (sphGFP and tpzGFP, respectively) that can be used to double-label cells. The system would then be set up by stable integration of a sphGFP encoding vector; introduction of an episomal vector encoding both HPRT1 and the tpzGFP; and, following retention or loss of this episomal survival plasmid, under different experimental conditions, by measuring the ratio between the fluorescent intensity of the two distinct GFPs over time.
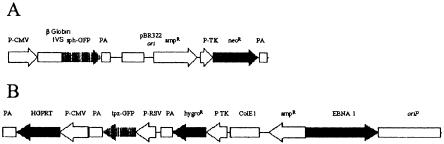
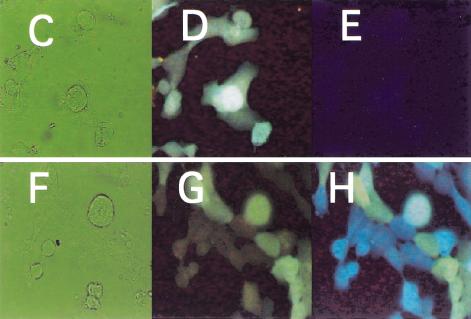
Figure 2A,B shows the respective plasmids that were prepared. As a recipient cell line, the HPRT1-deficient variant of the HT1080 fibrosarcoma cell line was chosen (Benedict et al. 1984). We generated cell clones that showed stable sphGFP expression. The HPRT1 survival plasmid was then introduced and a cell clone was chosen that stably expressed both the tpzGFP and the HPRT1 protein, as ascertained by resistance to HAT medium (Littlefield 1964). The fluorescence resulting from these two GFP variants, one stably integrated into the genome (sphGFP) and one episomal (tpzGFP), could be distinguished by use of two different microscope filter blocks (Fig. 2D,G vs. 2E,H).
Figure 2.
Structure and expression of the double-labeled GFP plasmid system. (A) The pIS integrating sphGFP vector. (B) The HPRT1-tpzGFP survival plasmid. P-CMV, P-TK, and P-RSV indicate the promoters of herpes TK1, CMV, and RSV, respectively. IVS represents the rabbit globin second intron. PA stands for polyadenylation signal. Solid arrows indicate open reading frames translated in the transduced human cells. (C–E) HIS4 cells expressing the pIS construct. (F–H) Clone 12 cells that express both the sphGFP from the pIS construct and tpzGFP from the episomal survival plasmid. Panels C and F are photographs of the phase image. Fluorescent images in panels D,E,F, and G were obtained using filter sets that preferentially detect sphGFP (D,G) or tpzGFP (E,H). The Clone 12 cells were maintained in hygromycin B selection to retain the survival plasmid.
These cells were then plated into 96-well microplates and scanned by a fluorescent microplate reader. Two separate filter sets could distinguish between the sphGFP and tpzGFP fluorescence. As expected, in the linear range of sphGFP reading a close correlation was seen between the number of cells and the fluorescent intensity (data not shown).
Fluorescent Measurement of Spontaneous Survival Plasmid Loss
We next examined whether spontaneous loss of the survival plasmid could be detected by fluorescence measurements. Since a high-throughput method necessitates use of microtiter plates, it was imperative to show that the expected plasmid loss occurs and could be detected in these growth conditions. However, the dynamics of cell division, and therefore the rate of the survival plasmid loss, could be very different in cells grown for long periods in microplates as opposed to cells stimulated to divide by a regimen of periodic trypsinization, dilution, and reseeding. Accordingly, we carried out measurement of fluorescence after removal of drug selection in cells continuously passaged in 90-mm plates as well as in cells grown for long periods in microtiter plates.
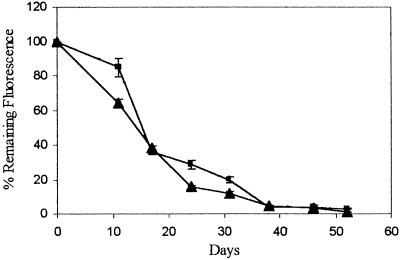
Results from one cell line, Clone 12, carrying an integrated sphGFP gene and an episome encoded tpzGFP are shown in Figure 3. Following removal of hygromycin B from the medium, the tpzGFP and sphGFP fluorescence ratios were monitored over time. The calculated ratio was normalized to readings taken at the same time points from cells kept continuously under hygromycin B selection. Fluorescence from the survival plasmid marked with tpzGFP decayed such that after ∼1 mo, Clone 12 lost 80%–90% of its initial fluorescence. Importantly, no significant difference in the rate of fluorescence decay could be detected between the cells maintained in microplate wells as opposed to those maintained by continuous passaging in petri dishes (Fig. 3).
Figure 3.
Spontaneous loss of tpzGFP-marked survival plasmid in Clone 12 cells. Cells were maintained in 96-well plates over the entire time period of the experiment (rectangles) or grown by continuous passaging in 90-mm petri dishes (triangles). Passaged cells were trypsinized and seeded into microplates on the day of reading. Plates were read using a microplate fluorescence reader. Data points show the mean relative fluorescence ratio between tpzGFP and sphGFP, expressed as a percentage of the same ratio measured from cells maintained under hygromycin B selection. All data represent the mean of fluorescence reading from at least three wells.
To test whether the gradual loss of tpzGFP fluorescence over time indeed reflects the loss of survival plasmid, two assays were conducted. In one, low molecular weight DNA present in Hirt supernatants (Hirt 1967) of Clone 12 cells was collected at various times after removal from hygromycin B selection. This DNA was used for bacterial transformation. We found that the decrease of ampicillin-resistant colonies correlated well with the loss of tpzGFP fluorescence over time (Table 1). In the second assay, a plasmid segregation assay (Reisman et al. 1985) was carried out. At each time point after drug removal, cells were reseeded into petri dishes containing hygromycin B. Our results show that the number of colonies able to grow in the presence of hygromycin B did indeed decrease at later time points (data not shown).
Table 1.
Monitoring of Spontaneous Survival Plasmid Loss by Hirt Supernatant Mediated Bacterial Transformation
| Days without selection | Bacterial transformant per 106 Clone 12 cells | Plasmid loss (k) per day (%)* |
|---|---|---|
| 0 | 628 | |
| 21 | 38 | 13.4 |
| 28 | 23 | 13.2 |
| 35 | 16 | 9.7 |
Clone 12 cells were plated at the beginning of the experiment in DMEM without hygromycin B. Cells were continuously passaged throughout the entire experiment. Low-molecular weight DNA present in Hirt supernatants was collected (Hirt 1967) at the indicated time points. All Hirt supernatants were normalized by addition of 1 ng of a chloramphenicol-resistant plasmid prior to the beginning of cell extraction. Each Hirt supernatant transformation of bacteria was plated both on chloramphenicol and ampicillin plates. Values in the table were normalized to the number of colonies counted on the chloramphenicol plates.
Calculated according to 2.303 log (N0/N) = kT, where N0 = % AmpR colonies at time zero (100%), N = % AmpR colonies remaining after propagation for T days without hygromycin B selection and k = % loss of plasmid per day.
Detection of Chemical-Induced Synthetic Lethality
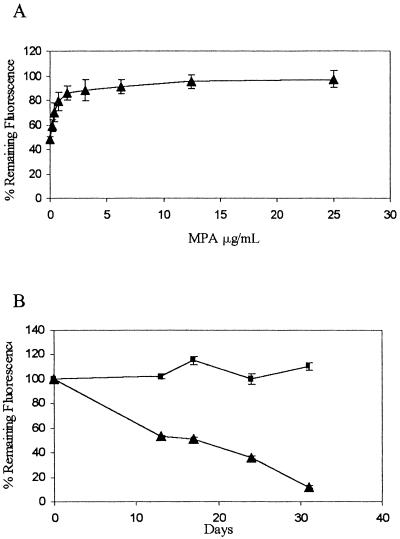
These results demonstrate that in the absence of selection, the HPRT1-tpzGFP survival plasmid is unstable in HPRT1-deficient HT1080 cells. Loss or retention of the plasmid can be determined by measuring the fluorescence intensity ratio in a microplate reader. We next tested whether these features would enable us to trace a synthetic lethal condition. The biosynthesis of GMP from IMP via XMP (xanthosine monophosphate) can be efficiently blocked using MPA, which inhibits IMPDH (Fig. 1). Under these conditions, normal HPRT1-positive cells survive by using guanine supplied in the medium to produce GMP via the salvage pathway, while HPRT1-deficient cells die. Clone 12, as an inherently HPRT1-deficient cell line, must retain the HPRT1-tpzGFP survival plasmid to stay alive in this synthetic lethal situation. Clone 12 cells grown in medium supplemented with guanine spontaneously lost the survival plasmid (Fig. 4A). In contrast, addition of MPA at successively higher concentrations to medium supplemented with guanine caused increasing retention of the survival plasmid, which could be detected by an increase in the tpzGFP to sphGFP fluorescence ratio (Fig. 4A). We observed a dose response which reached a plateau at fluorescence ratio levels similar to those obtained when hygromycin B selection was imposed (Fig. 4B). Moreover, as shown in Figure 4B, when fluorescence was observed over time, MPA could cause retention of the survival plasmid for the entire time period, while cells without MPA continued to lose tpzGFP fluorescence.
Figure 4.
Synthetic lethality imposed by MPA causes retention of the survival plasmid. (A) Cells were maintained in 96-well microplates and hygromycin B was removed and replaced with medium containing 35 μM guanine and MPA in serial dilutions starting with 25 μg/mL. Microplates were read after 2 wk. (B) Cells in 96-well microplates were maintained over the entire time period in either medium with 35 μM guanine and 12.5 μg/mL MPA (rectangles) or in guanine-supplemented medium alone (triangles). Plate reading and data representation are as in Fig. 3.
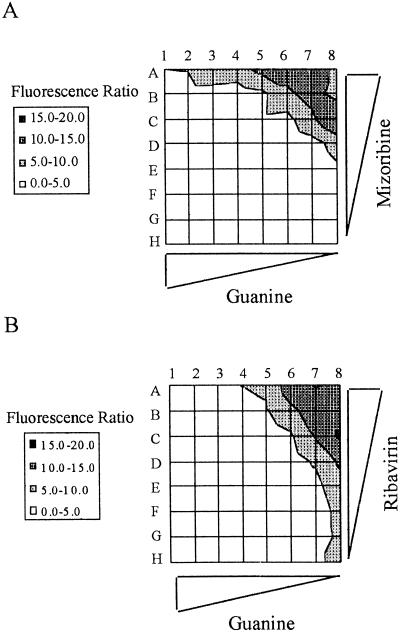
IMPDH has two isoforms and is considered to be the rate-limiting enzyme in guanine nucleotide biosynthesis (Haste et al. 1999). Two basic types of drugs can effectively inhibit the enzyme: nucleoside analogs and nonnucleoside inhibitors. MPA is of the second class and binds the NAD site within the enzyme. Nucleoside analog inhibitors bind to the IMP substrate site (Haste et al. 1999). Accordingly, we asked whether we could detect synthetic lethality when nucleoside analog inhibitors were applied to Clone 12 cells. We chose for this assay two drugs, ribavirin and mizoribine, both in use against viral infections. We tested both drugs on Clone 12 cells grown in 96-well microtiter plates in serial dilutions of the drugs as well as serial dilutions of guanine. The observed matrices of results are shown in Figure 5. Both drugs caused retention of the survival plasmid, in a way that was dependent on the concentration of each drug as well as that of guanine. These matrices allowed sensitive measurement of the synthetic lethal effects imposed by these nucleoside analogs.
Figure 5.
Chemical synthetic lethality induced by nucleoside analog inhibitors of IMPDH. Clone 12 cells were grown in microplates in the presence of mizoribine (A) or ribavirin (B) for a period of 3 wk before measurement of fluorescent ratio. Values for the fluorescent ratios are shown on the left. Rows A–H, in each matrix, represent the inhibitory drug data. Guanine was at 400 μM in column 1, and serial dilutions were in the indicated direction. In (A), mizoribine was at 400 μg/mL in row A and serial dilutions were in the indicated direction. In (B), ribavirin was at 200 μg/mL in row A and serial dilutions were in the indicated direction.
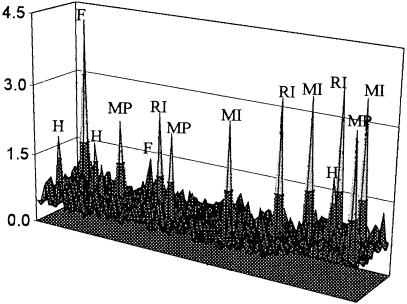
Next, we tested if the synthetic lethality assay could be used in a blind test to screen for these IMPDH inhibitors. Clone 12 was seeded into 1200 wells in microtiter plates in medium lacking selection. MPA, ribavirin, mizoribine, and hygromycin B were added at random (together with guanine) to three wells. Alanosine, an inhibitor of adenylosuccinate synthase (an enzyme in the pathway converting IMP to AMP) was also added as a negative control. As expected, the survival plasmid was not retained in the presence of alanosine (data not shown). However, the presence of all three IMPDH inhibitors and hygromycin B was clearly detected, as were two wells containing false-positive cells (Fig. 6). The calculated false positive rate for this experiment was 1/600 and was similar to previous control experiments (data not shown).
Figure 6.
Detection of chemical synthetic lethality in a blind large-scale test. Clone 12 cells were seeded into 1200 wells in 96-well microplates. Five chemicals were randomly added to three wells each; hygromycin B (H) at 200 μg/mL, MPA (MP) at 25 μg/mL, ribavirin (RI) at 50 μg/mL, mizoribine (MI) at 200 μg/mL, and alanosine at 10 μg/mL. MPA was added in the presence of 200 μM guanine, ribavirin, and mizoribine in 12.5 μM guanine, while alanosine was added together with 60 μM adenine. Plates were read after 28 d, and fluorescent ratios are displayed as in Fig. 5. False-positive wells (F) are also labeled.
DISCUSSION
In this article, we demonstrate the feasibility of a chemical synthetic lethality screen in cultured human cells, using a sensitive fluorescent assay. Imposition of synthetic lethality by the addition of the IMPDH inhibitors MPA, ribavirin, and mizoribine prevented the spontaneous loss of the survival plasmid. The retention or loss of the plasmid was sensitively measured over time, using a microplate fluorescence reader while cells were grown in microtiter plates. The surprising sensitivity of this assay was such that small changes in the amount of chemical inhibitor or metabolites in the medium were easily detected.
We have performed this cell-based screen in several subclones of the parental variant HT1080 cell line, as well as in the HPRT-deficient LNSV human cell line (data not shown). This demonstrates the generality of this assay in cultured human cells and that the results are not specific to Clone 12.
The ability to elucidate the interaction between specific chemical reagents and a single gene, when linked to high-throughput scanning methodology, should be very useful in the search for drugs that synergize with particular gene deficiencies to cause cell lethality. In particular, as the assay is based on identification of a lethal phenotype, one could scan a library of small organic chemical compounds (Mitchison 1994) or antisense phosphorothioate oligonucleotides (Agrawal and Zhao 1998) for reagents that kill either a benign or cancerous cell in a defined genetic milieu, where the reagent is synthetic lethal with a defined mutant gene. This possibility was demonstrated, albeit on a relatively small scale, in the experiment described in Figure 6. Notably, this system could also expedite the search for better IMPDH inhibitors, a goal that remains relevant for the finding of new immunosuppressive, antiviral, and antitumor agents (Franklin et al. 1999). The chemical synthetic lethality assay in its current form requires a relatively extended period of time for appreciable loss of the survival plasmid. Monitoring of cell response to a drug over time is in marked contrast to short-term assays such as those performed in the extensive NCI compound screens (Weinstein et al. 1997). This may actually prove to be beneficial as short-term assays overlook slow death phenomena that are nonapoptotic for the most part.
Following discovery of chemicals that are synthetically lethal with the gene of interest, the task of identification of the drug target may be achieved by several approaches. One method that is particularly relevant to the chemical synthetic lethality screen would be to introduce a human CDNA library, on an episomal vector, into the parental cell line that has been used in the assay. One may assume that in the absence of the survival plasmid, any drug selected in the screen is lethal to the parental cell line, which is deficient in the gene of interest. Excess expression of the protein that is affected by the drug would allow the respectively transfected cell to survive in the presence of the drug, while other cells would die. The episomal vector encoding the putative drug target would then be rescued from the colonies able to grow in the presence of the minimal lethal dose of the drug. Another emerging avenue of drug target identification is inspection of whole-genome changes in RNA expression that result from growth in the presence of the drug, using DNA microarrays (Marton et al. 1998; Debouck and Metcalf 2000). This should be eventually aided by comparison to a predetermined reference database or compendium of whole-genome expression profiles of known mutations and drugs with known molecular targets (Hughes et al. 2000). Also, one may be able to examine direct drug binding to polypeptide targets in protein microarrays (MacBeath and Schreiber 2000) or drug-mediated ablation of protein–protein interactions in a two-hybrid system (Vidal and Endoh 1999).
The current method has a potential genetic synthetic lethality aspect, in that antisense oligonucleotides can be applied to cells much as chemical reagents are. Further extension of this potential could also be achieved through expression of libraries of antisense RNA (Deiss and Kimchi 1991; Gudkov et al. 1993), truncated cDNA (Gudkov et al. 1993), ribozymes (Breaker and Joyce 1994), RNA aptamers (Thomas et al. 1997), or peptide aptamers (Colas et al. 1996; Geyer et al. 1999). The sorting of the dominant genetic element(s) within any of these libraries would require enrichment steps, such as FACS, coupled with high-throughput screening.
The synthetic lethality screen has been particularly useful in yeast research, where gene knockouts can be easily obtained by homologous recombination. The availability of a large number of mutant human cell lines derived from genetic disorders on the one hand and continuing research into aberrant gene expression in different cell lines on the other should enrich the growing reservoir of potential recipient cells and genes of interest. Also, new and better technologies are evolving that employ homologous recombination for gene disruption in somatic human cells (Chan et al. 1999; Sedivy et al. 1999). Taking into account the genetic differences between established cell lines and tumors, cross-reference with the genetic molecular characterization of a given cancer obtained via the Cancer Genome Anatomy Project (Strausberg et al. 2000), and other sources will be important.
Implementation of any of the dominant-negative strategies mentioned above should allow for the identification of a human gene(s) synthetically lethal with a human gene of interest. Such genetic synthetic lethality screens should have a major impact on human functional genomics, as well as identification of target genes for cancer therapy.
It is important to mention a complementary genetic approach, the Seattle Project (Hartwell 1997), in which screening for new cancer drugs or identification of new cancer drug targets is initiated by studies performed in yeast. The rationale behind this work is that several basic mechanisms governing genomic stability, and thus cancer susceptibility, have been conserved from model organisms such as yeast, nematodes, and fruit flies to humans. Accordingly, some of the alterations found in human tumors can be modeled in these simpler organisms that are more amenable to genetic manipulation. Consequently, yeast mutants are currently being screened for either chemicals with a preferred therapeutic advantage or second-site mutations that are lethal only in the mutant yeast strain (Hartwell et al. 1997). The latter is being performed by synthetic lethality screen in yeast (Bender and Pringle 1991). Either one of these elegant strategies requires in its second phase the uncertain and complicated extrapolation back to human tumors, as discussed (Hartwell et al. 1997). The proposed chemical and genetic synthetic lethality screens in cultured human cells are technically difficult to pursue in comparison to the yeast system. Nevertheless, they have the significant advantage of directly addressing human tumor cell lines.
METHODS
Construction of Plasmids
pIS was constructed by replacing the BamHI fragment encoding CD20 from pCMV-CD20 (kind gift from S. van den Heuvel and E. Harlow, M.G.H. Cancer Center) with a blunt-ended HindIII-BamHI fragment containing the coding sequence of sphGFP from the pGFPsph-b [R] vector (Packard Instruments). The episomal HPRT1-tpzGFP survival plasmid was constructed by first cloning a HindIII-BamHI blunt-ended fragment encoding the tpzGFP and polyadenylation signal from the pGFPtpz-b [R] vector (Packard Instruments) into the HindIII site of pCEP4 (Invitrogen). The coding sequence of HPRT1 was cloned into pcDNA3 (Invitrogen) and subsequently removed together with the CMV promoter by digestion with BglII and BamHI. This fragment was then cloned into the BamHI site of the pCEP4-GFP vector. The final survival plasmid was produced by cloning the PvuII-XbaI fragment of the pCEP4-HPRT1-tpzGFP vector into the PvuII-XbaI site of pREP4 (Invitrogen), so that transcription of the tpzGFP gene is initiated from the RSV promoter.
Expression of Constructs in Cells
HPRT1-deficient HT1080 fibrosarcoma cells (kind gift from A. Ganesan, Stanford University; Benedict et al. 1984) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 4 mM L-glutamine. Transfections were carried out using the calcium phosphate precipitation technique as previously described (Teitz et al. 1987). For pCMV-GFP, selection in G418 (Calbiochem) was carried out at 400 μg/mL, while maintenance was at 50 μg/mL. For the HPRT1-tpzGFP survival plasmid, selection in hygromycin B (Sigma) was at 150 μg/mL, while maintenance was at 50 μg/mL. Selection and maintenance of the survival plasmid was also carried out in HAT medium (100 μM hypoxanthine, 0.4 μM aminopterin, 16 μM thymidine; Littlefield 1964). MPA was obtained from Eli Lily, while ribavirin and mizoribine were obtained from Sigma.
Microscopic images were captured using an Olympus BX40 microscope with a Sony CCD-Iris color video camera. SphGFP was visualized using a filter block with an excitation bandpass of 340–380 nm. TpzGFP was visualized using a filter block with an excitation bandpass of 450–490 nm.
Fluorescent Scanning of Microtiter Plates
For fluorescent scanning, cells were trypsinized and distributed at 30,000 cells/well into 96-well microplates (TPP). Growth medium was changed twice a week, and plates were maintained for up to 75 d. Cells remained viable over the entire time period. Plates were prepared for scanning by replacement of medium in wells with Hank's balanced salt solution without phenol red. This procedure greatly minimized background fluorescence from the growth medium while maintaining maximal viability. Plates were scanned with an FL600 microplate fluorescence reader using the KC4 software (Biotek Instruments). Excitation for sphGFP was at 380 nm with a bandpass of 20 nm, while emission was measured at 508 nm with a bandpass of 40 nm. Excitation of tpzGFP was at 485 nm with a bandpass of 20 nm, while emission was measured at 535 nm with a bandpass of 40 nm. To avoid possible artifacts, all wells within a given experiment were assayed for the fluorescence of the two GFP mutants using fixed sensitivities. Integrated sphGFP was used as an internal control for the number of cells. This was achieved by dividing the relative fluorescence resulting from the episomal tpzGFP vector by the relative fluorescence for sphGFP for each well. This ratio was then divided by the average fluorescence ratio for cells maintained under hygromycin B or HAT selection, resulting in a value representing percentage remaining fluorescence for each well as compared with wells maintained under continuous selection. The data points are an average for all wells and the calculated standard deviation. Cells were returned to growth medium immediately following scanning.
Assessment of Episomal Survival Plasmid Levels
To assay for β-lactamase gene expression, low–molecular weight DNA was extracted according to Hirt 1967 and transformed into DH10B bacteria by electroporation, followed by selection for ampicillin resistance. Expression of the hygromycin B resistance gene from the survival plasmid was monitored by the plasmid segregation assay (Reisman et al. 1985).
Acknowledgments
We thank A. Ganesan, T. Yang, S. van der Heuvel, and E. Harlow for reagents and I. Tzarfati for his help in photography. We also thank H. Cedar and M. Revel for critical reading of this manuscript. Special thanks are due to W. Richardson (Packard Instruments) for his kind assistance in early obtainment of the GFP mutant genes. This work was supported by a biotechnological infrastructure grant in the field of functional genomics to D.C. and Y.O. from the Israeli Ministry of Science and Technology.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL canaani@post.tau.ac.il; FAX: 972-3-6424270.
Article and publication are at www.genome.org/cgi/doi/10.1101/gr.154201.
REFERENCES
- Agrawal S, Zhao Q. Antisense therapeutics. Curr Opin Chem Biol. 1998;2:519–528. doi: 10.1016/s1367-5931(98)80129-4. [DOI] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict WF, Weissman BE, Mark C, Stanbridge EJ. Tumorigenicity of human HT1080 fibrosarcoma × normal fibroblast hybrids: Chromosome dosage dependency. Cancer Res. 1984;44:3471–3479. [PubMed] [Google Scholar]
- Breaker RR, Joyce GF. Inventing and improving ribozyme function: Rational design versus iterative selection methods. Trends Biotechnol. 1994;12:268–275. doi: 10.1016/0167-7799(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Calos MP. The potential of extrachromosomal replicating vectors for gene therapy. Trends Genet. 1996;12:463–466. doi: 10.1016/0168-9525(96)40049-x. [DOI] [PubMed] [Google Scholar]
- Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14–3–3 ς is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature. 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- Debouck C, Metcalf B. The impact of genomics on drug discovery. Annu Rev Pharmacol Toxicol. 2000;40:193–207. doi: 10.1146/annurev.pharmtox.40.1.193. [DOI] [PubMed] [Google Scholar]
- Deiss LP, Kimchi A. A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science. 1991;252:117–120. doi: 10.1126/science.1901424. [DOI] [PubMed] [Google Scholar]
- Fields S. The future is function. Nat Genet. 1997;15:325–327. doi: 10.1038/ng0497-325. [DOI] [PubMed] [Google Scholar]
- Franklin TJ, Morris WP, Jacobs VN, Culbert EJ, Heys CA, Ward WH, Cook PN, Jung F, Ple P. A novel series of non-nucleoside inhibitors of inosine 5′-monophosphate dehydrogenase with immunosuppressive activity. Biochem Pharmacol. 1999;58:867–876. doi: 10.1016/s0006-2952(99)00170-7. [DOI] [PubMed] [Google Scholar]
- Geyer CR, Colman-Lerner A, Brent R. “Mutagenesis” by peptide aptamers identifies genetic network members and pathway connections. Proc Natl Acad Sci. 1999;96:8567–8572. doi: 10.1073/pnas.96.15.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudkov AV, Zelnick CR, Kazarov AR, Thimmapaya R, Suttle DP, Beck WT, Roninson IB. Isolation of genetic suppressor elements, inducing resistance to topoisomerase II-interactive cytotoxic drugs, from human topoisomerase II cDNA. Proc Natl Acad Sci. 1993;90:3231–3235. doi: 10.1073/pnas.90.8.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- Haste S, De Clerq E, Balzarini T. Role of antimetabolites of purine and pyrimidine nucleotide metabolism in tumor cell differentiation. Biochem Pharmacol. 1999;58:539–555. doi: 10.1016/s0006-2952(99)00035-0. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennet HA, Coffey E, Dai H, He YD, et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Lander E. The new genomics: Global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- Littlefield JW. Selection of hybrids from matings of fibroblasts in vitro and their presumed recombinants. Science. 1964;145:709–710. doi: 10.1126/science.145.3633.709. [DOI] [PubMed] [Google Scholar]
- MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts CJ, Stoughton R, Burchard J, Slade D, Dai H, et al. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. Towards a pharmacological genetics. Chem Biol. 1994;1:3–6. doi: 10.1016/1074-5521(94)90034-5. [DOI] [PubMed] [Google Scholar]
- Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedivy JM, Vogelstein B, Liber HL, Hendrickson EA, Rosmarin A. Gene targeting in human cells without isogenic DNA. Science. 1999;283:9. [Google Scholar]
- Strausberg RL, Buetow KH, Emmert-Buck MR, Klausner RD. The cancer genome anatomy project: Building an annotated gene index. Trends Genet. 2000;16:103–106. doi: 10.1016/s0168-9525(99)01937-x. [DOI] [PubMed] [Google Scholar]
- Teitz T, Naiman T, Avissar SS, Bar S, Okayama H, Canaani D. Complementation of the UV-sensitive phenotype of a xeroderma pigmentosum human cell line by transfection with a cDNA clone library. Proc Natl Acad Sci. 1987;84:8801–8804. doi: 10.1073/pnas.84.24.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitz T, Naiman T, Eli D, Bakhanashvili M, Canaani D. Complementation of excision-repair deficiency in a human cell: Advantage in the use of a cDNA clone library for gene transfer. In: Beaudet AL, et al., editors. Gene transfer and gene therapy. 1989. , UCLA symposia on molecular and cellular biology. Vol. 87, pp. 215–223. Liss, New York. [Google Scholar]
- Teitz T, Eli D, Penner M, Bakhanashvili M, Naiman T, Timme TL, Wood CM, Moses RE, Canaani D. Expression of the cDNA for the beta subunit of human casein kinase II confers partial UV resistance on xeroderma pigmentosum cells. Mutat Res. 1990;236:85–97. doi: 10.1016/0921-8777(90)90036-5. [DOI] [PubMed] [Google Scholar]
- Thomas M, Chedin S, Carles C, Riva M, Famulok M, Sentenac A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J Biol Chem. 1997;272:27980–27986. doi: 10.1074/jbc.272.44.27980. [DOI] [PubMed] [Google Scholar]
- Vidal M, Endoh H. Prospects for drug screening using the reverse two-hybrid system. Trends Biotechnol. 1999;17:374–381. doi: 10.1016/s0167-7799(99)01338-4. [DOI] [PubMed] [Google Scholar]
- Weinstein JN, Myers TG, O'Connor PM, Friend SH, Fornace AJ, Jr, Kohn KW, Fojo T, Bates SE, Rubinstein LV, Anderson NL, et al. An information-intensive approach to the molecular pharmacology of cancer. Science. 1997;275:343–349. doi: 10.1126/science.275.5298.343. [DOI] [PubMed] [Google Scholar]
- Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]