Abstract
Background
Sirtuin is a member of the nicotinamide adenine dinucleotide (NAD)-dependent deacetylases, and has been reported to play a pivotal role in energy expenditure, mitochondrial function and pathogenesis of metabolic diseases, including aging kidneys. In this study, we focused on the genes encoding sirtuin families, and examined the association between single nucleotide polymorphisms (SNPs) within genes encoding sirtuin families and diabetic nephropathy.
Methods
We examined 52 SNPs within the SIRT genes (11 in SIRT1, 7 in SIRT2, 14 in SIRT3, 7 in SIRT4, 9 in SIRT5, and 4 in SIRT6) in 3 independent Japanese populations with type 2 diabetes (study 1: 747 cases (overt proteinuria), 557 controls; study 2: 455 cases (overt proteinuria) and 965 controls; study 3: 300 cases (end-stage renal disease) and 218 controls). The associations between these SNPs were analyzed by the Cochran–Armitage trend test, and results of the 3 studies were combined with a meta-analysis. We further examined an independent cohort (195 proteinuria cases and 264 controls) for validation of the original association.
Results
We identified 4 SNPs in SIRT1 that were nominally associated with diabetic nephropathy (P < 0.05), and subsequent haplotype analysis revealed that a haplotype consisting of the 11 SNPs within SIRT1 locus had a stronger association (P = 0.0028).
Conclusion
These results indicate that SIRT1 may play a role in susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes.
Electronic supplementary material
The online version of this article (doi:10.1007/s10157-011-0418-0) contains supplementary material, which is available to authorized users.
Keywords: Single nucleotide polymorphism (SNP), Association study, SIRT1, Diabetic nephropathy
Introduction
Diabetic nephropathy is a serious microvascular complication of diabetes, and is a leading cause of end-stage renal disease in Western countries [1] and in Japan [2]. The escalating prevalence and limitation of currently available therapeutic options highlight the need for a more accurate understanding of the pathogenesis of diabetic nephropathy. Several environmental factors, such as medication, daily energy consumptions, and daily sodium intake, are likely to cooperate with genetic factors to contribute to its development and progression [3, 4]; however, the precise mechanism for this contribution is unknown. Krolewski et al. [5] reported that the cumulative incidence of diabetic retinopathy increased linearly with duration of diabetes, whereas the occurrence of nephropathy was almost none after 20–25 years of diabetes duration, and only a modest number of individuals with diabetes (~30%) developed diabetic nephropathy. Familial clustering of diabetic nephropathy was also reported in both type 1 [4] and type 2 diabetes [6]; thus, the involvement of genetic factors in the development of diabetic nephropathy is strongly suggested. Both candidate gene approaches and genome-wide linkage analyses have suggested several candidate genes with a potential impact on diabetic nephropathy. These findings, however, have not been robustly replicated and many genes responsible for susceptibility to diabetic nephropathy remain to be identified. To identify loci involved in susceptibility to common diseases, we initiated the first round of a genome-wide association study (GWAS) using 100,000 single nucleotide polymorphisms (SNPs) from a Japanese SNP database (JSNP: http://snp.ims.u-tokyo.ac.jp/index_ja.html). Through this project, we have previously identified genes encoding solute carrier family 12 (sodium/chloride) member 3 (SLC12A3, MIM 600968, Online Mendelian Inheritance in Man: http://www.ncbi.nlm.nih.gov/omim) [7]; engulfment and cell motility 1 (ELMO1, MIM 606420) [8]; neurocalcin δ (NCALD, MIM 606722) [9]; and acetyl-coenzyme A carboxylase beta gene (ACACB, MIM: 601557) [10] as being associated with susceptibility to diabetic nephropathy. The association between ELMO1 or ACACB and diabetic nephropathy has been confirmed in different ethnic populations [11–13]. The GWAS for diabetic nephropathy using European American populations (the Genetics of Kidneys in Diabetes (GoKinD) collection) led to the identification of 4 distinct loci as novel candidate loci for susceptibility to diabetic nephropathy in European American subjects with type 1 diabetes [14]: the CPVL/CHN2 locus on chromosome 7, the FRMD3 locus on chromosome 9, the CARS locus on chromosome 11, and a locus near IRS2 on chromosome 13. Among those 4 loci, only one locus (near IRS2 in chromosome 13) could be replicated in Japanese subjects with type 2 diabetes [15]. Although these loci are considered as convincing susceptibility loci for diabetic nephropathy across different ethnic groups, a considerable number of susceptibility genes for diabetic nephropathy still remain to be identified.
Sirtuins, the silent information regulator-2 (SIR2) family, is a member of NAD-dependent deacetylases, and the sir2 gene was originally identified as a gene affecting the malting ability of yeast. Mammalian sirtuins consist of seven members, SIRT1–SIRT7, and some of them, especially SIRT1, have been shown to play pivotal roles in the regulation of aging, longevity, or in the pathogenesis of age-related metabolic diseases, such as type 2 diabetes [16–18]. The expressions of sirtuin families have also been observed in the kidneys, and recently SIRT1 has been shown to mediate a protective role of calorie restriction (CR) in the progression of the aging kidney [19]. These observations suggest the possibility that mammalian sirtuins are a candidate for conferring susceptibility to diabetic nephropathy.
In order to test this hypothesis, we focused on genes encoding mammalian sirtuins as candidate genes for diabetic nephropathy and investigated the association between SNPs within the SIRT genes and diabetic nephropathy in Japanese subjects with type 2 diabetes.
Materials and methods
Subjects, DNA preparation
Study 1
DNA samples were obtained from the peripheral blood of patients with type 2 diabetes who regularly visited the outpatient clinic at Shiga University of Medical Science, Tokyo Women’s Medical University, Juntendo University, Kawasaki Medical School, Iwate Medical University, Toride Kyodo Hospital, Kawai Clinic, Osaka City General Hospital, Chiba Tokushukai Hospital, or Osaka Rosai Hospital. Diabetes was diagnosed according to the World Health Organization criteria. Type 2 diabetes was clinically defined as a disease with gradual adult onset. Subjects who tested positive for anti-glutamic acid decarboxylase antibodies and those diagnosed with mitochondrial disease (mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS)) or maturity onset diabetes of the young were not included. The patients were divided into 2 groups: (1) the nephropathy group (n = 754, age 60.1 ± 0.4, diabetes duration 19.3 ± 0.4, body mass index (BMI) 23.7 ± 0.2, mean ± SE) comprised patients with diabetic retinopathy and overt nephropathy indicated by a urinary albumin excretion rate (AER) ≥200 μg/min or a urinary albumin/creatinine ratio (ACR) ≥300 mg/g creatinine (Cr), and (2) the control group (n = 558, age 62.4 ± 0.5, diabetes duration 15.3 ± 0.4, BMI 23.6 ± 0.2) comprised patients who had diabetic retinopathy but no evidence of renal dysfunction (i.e. AER <20 μg/min or ACR <30 mg/g Cr). The AER or ACR were measured at least twice for each patient.
Study 2
We selected diabetic nephropathy patients and control patients among the subjects enrolled in the BioBank Japan. Nephropathy cases were defined as patients with type 2 diabetes having both overt diabetic nephropathy and diabetic retinopathy (n = 449, age 64.7 ± 0.4, BMI 23.5 ± 0.2). The control subjects were patients with type 2 diabetes who had diabetic retinopathy and normoalbuminuria (n = 965, age 64.8 ± 0.3, BMI 23.8 ± 0.1).
Study 3
Patients with type 2 diabetes who regularly visited Tokai University Hospital or its affiliated hospitals were enrolled in this study. All the nephropathy patients (n = 300, age 64.4 ± 0.6, diabetes duration 21.9 ± 0.9, BMI 22.1 ± 0.2, mean ± SE) were receiving chronic hemodialysis therapy, and the control patients (n = 224, age 65.0 ± 0.7, diabetes duration 16.3 ± 0.4, BMI 23.4 ± 0.3, mean ± SE) included those with normoalbuminuria as determined by at least 2 measurements of urinary ACR and with diabetes for >10 years.
All the patients participating in this study provided written informed consent, and the study protocol was approved by the ethics committees of RIKEN Yokohama Institute and of each participating institution.
The clinical characteristics of the subjects are shown in Supplementary Table 1.
SNP genotyping
We searched the HapMap database (http://hapmap.ncbi.nlm.nih.gov/) for SNPs within the genes encoding sirtuin families, and selected 55 SNPs (39 tagging SNPs) for genotyping; 11 in SIRT1 (rs12778366, rs3740051, rs2236318, rs2236319, rs10823108, rs10997868, rs2273773, rs3818292, rs3818291, rs4746720, rs10823116), 7 in SIRT2 (rs1001413, rs892034, rs2015, rs2241703, rs2082435, rs11575003, rs2053071), 15 in SIRT3 (rs11246002, rs2293168, rs3216, rs10081, rs511744, rs6598074, rs4758633, rs11246007, rs3782117, rs3782116, rs3782115, rs1023430, rs12576565, rs536715, rs3829998), 7 in SIRT4 (rs6490288, rs7298516, rs3847968, rs12424555, rs7137625, rs2261612, rs2070873), 11 in SIRT5 (rs2804923, rs9382227, rs2804916, rs2804918, rs9370232, rs4712047, rs3734674, rs11751539, rs3757261, rs2253217, rs2841514), and 4 in SIRT6 (rs350852, rs7246235, rs107251, rs350844). We could not identify any confirmed SNPs within SIRT7 in the Japanese population. The genotyping of these SNPs was performed by using multiplex polymerase chain reaction (PCR)-invader assays, as described previously [7–10].
Statistical analyses
We tested the genotype distributions for Hardy–Weinberg equilibrium (HWE) proportions by using the chi-squared test. We analyzed the differences between the case−control groups in terms of the distribution of genotypes with the Cochran–Armitage trend test. The analyses for haplotype structures within each gene were performed using Haploview software version 4.1 [20]. A combined meta-analysis was performed using the Mantel–Haenszel procedure with a fixed effects model after testing for heterogeneity.
Results
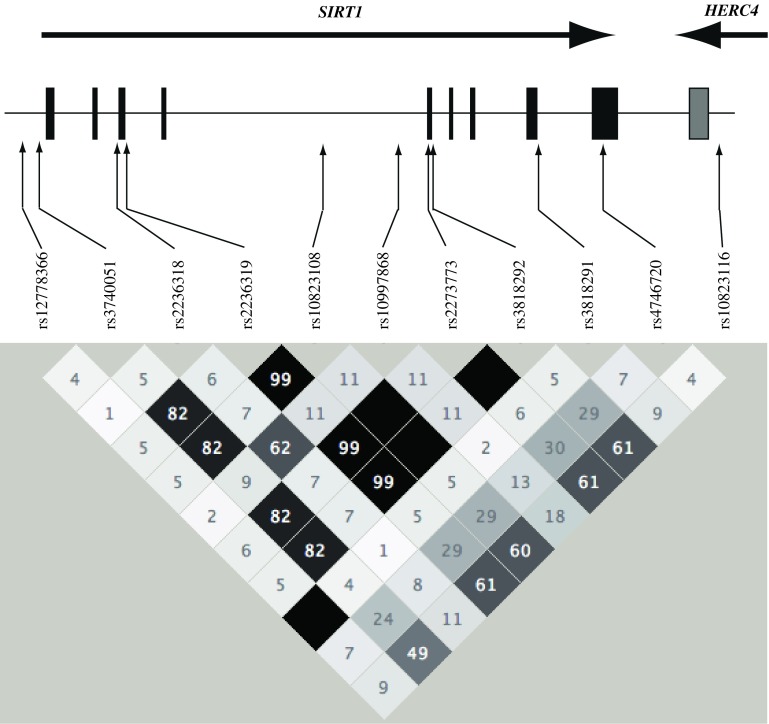
Among the 55 SNPs examined, genotype distributions of 3 SNPs, rs12576565 in SIRT3, and rs2804923 and rs2841514 in SIRT 5, showed significant deviation from HWE proportion in control groups (P < 0.01, Supplementary Table 2), and these 3 SNPs were excluded from the association study. As shown in Table 1, 8 out of 11 SNPs in SIRT1 showed a directionally consistent association with diabetic nephropathy in all 3 studies, although individual associations were not significant (P > 0.05, Supplementary Table 2). In a combined meta-analysis, we could identify a nominally significant association between rs4746720 and proteinuria, and between 4 SNPs, rs2236319, rs10823108, rs3818292, rs4746720, and combined phenotypes (proteinuria + ESRD, P < 0.05). Subsequent haplotype analysis revealed that the 11 SNPs formed one haplotype block (Fig. 1), and 7 common haplotypes covered >99% of the present Japanese population. Among them one haplotype had a stronger association with diabetic nephropathy than single SNPs alone (P = 0.016, odds ratio (OR) 1.31 95% confidence interval (CI) 1.05–1.62]. Any SNPs or haplotypes in SIRT2–6 were not associated with diabetic nephropathy in the combined analysis (Tables 2, 3, 4, 5, 6), although there was an association between 3 SNPs (rs4712047, rs3734674, rs3757261) in SIRT5 and diabetic nephropathy in the study 2 population (Supplementary Table 2). To validate the association between SIRT1 and diabetic nephropathy, we examined another 195 cases (overt proteinuria) and 264 controls registered in the BioBank Japan (study 4). As shown in Table 7, most SNPs showed a consistent association with those in the original finding, and the association of the haplotype was strengthened further (P = 0.0028, OR 1.36, 95% CI 1.11–1.66). We further examined the association between SIRT1 SNPs and microalbuminuria in studies 1 and 2, but could not identify a significant association (Supplementary Table 3), suggesting SIRT1 SNPs might contribute to the progression of nephropathy rather than its onset in patients with type 2 diabetes.
Table 1.
Association between SNPs in SIRT1 and diabetic nephropathy
| Allele frequencies (nephropathy case−control) | |||||||
|---|---|---|---|---|---|---|---|
| Proteinuria | ESRD | Combined | |||||
| Study 1 | Study 2 | P | OR (95% CI) | Study 3 | P | OR (95% CI) | |
| SNP | |||||||
| rs12778366a T>C | 0.111/0.103 | 0.125/0.124 | 0.672 | 1.04 (0.86–1.26) | 0.101/0.119 | 0.981 | 0.998 (0.84–1.18) |
| rs3740051a A>G | 0.291/0.277 | 0.316/0.301 | 0.299 | 1.07 (0.94–1.22) | 0.310/0.274 | 0.138 | 1.09 (0.97–1.23) |
| rs2236318a T>A | 0.121/0.129 | 0.099/0.111 | 0.327 | 0.91 (0.75–1.10) | 0.106/0.119 | 0.236 | 0.90 (0.76–1.07) |
| rs2236319 A>G | 0.339/0.317 | 0.358/0.339 | 0.165 | 1.09 (0.96–1.24) | 0.349/0.300 | 0.048 | 1.12 (1.00–1.26) |
| rs10823108 G>A | 0.335/0.318 | 0.357/0.335 | 0.169 | 1.09 (0.96–1.24) | 0.351/0.302 | 0.049 | 1.12 (1.00–1.26) |
| rs10997868a C>A | 0.187/0.184 | 0.187/0.174 | 0.520 | 1.05 (0.90–1.23) | 0.180/0.173 | 0.482 | 1.05 (0.91–1.21) |
| rs2273773 T>C | 0.339/0.325 | 0.361/0.347 | 0.325 | 1.07 (0.94–1.21) | 0.353/0.306 | 0.113 | 1.10 (0.98–1.23) |
| rs3818292 A>G | 0.336/0.317 | 0.360/0.335 | 0.134 | 1.10 (0.97–1.25) | 0.352/0.306 | 0.042 | 1.13 (1.00–1.26) |
| rs3818291 G>A | 0.111/0.101 | 0.127/0.129 | 0.650 | 1.04 (0.87–1.26) | 0.101/0.124 | 0.927 | 0.99 (0.84–1.17) |
| rs4746720a T>C | 0.366/0.394 | 0.331/0.364 | 0.041 | 0.88 (0.77–0.99) | 0.367/0.400 | 0.021 | 0.88 (0.78–0.98) |
| rs10823116a A>G | 0.446/0.442 | 0.441/0.448 | 0.905 | 0.99 (0.88–1.12) | 0.459/0.394 | 0.428 | 1.05 (0.94–1.16) |
| Haplotype | |||||||
| TGTGACCGGTG | 0.294/0.279 | 0.316/0.300 | 0.250 | 1.08 (0.95–1.23) | 0.315/0.273 | 0.095 | 1.10 (0.98–1.24) |
| TATAGCTAGCA | 0.255/0.273 | 0.251/0.252 | 0.464 | 0.95 (0.83–1.09) | 0.253/0.304 | 0.143 | 0.91 (0.81–1.03) |
| CATAGCTAATA | 0.112/0.103 | 0.124/0.129 | 0.817 | 1.02 (0.85–1.23) | 0.100/0.119 | 0.841 | 0.98 (0.83–1.16) |
| TAAAGATAGTA | 0.123/0.128 | 0.104/0.112 | 0.484 | 0.94 (0.78–1.13) | 0.105/0.122 | 0.319 | 0.92 (0.78–1.08) |
| TATAGCTAGCG | 0.109/0.123 | 0.085/0.111 | 0.037 | 0.81 (0.67–0.99) | 0.113/0.099 | 0.117 | 0.87 (0.73–1.03) |
| TATAGATAGTA | 0.065/0.055 | 0.078/0.059 | 0.051 | 1.27 (0.998–1.61) | 0.077/0.053 | 0.016 | 1.31 (1.05–1.62) |
| TATGACCGGTG | 0.042/0.039 | 0.040/0.036 | 0.57 | 1.09 (0.81–1.48) | 0.036/0.028 | 0.421 | 1.12 (0.85–1.48) |
aTag SNPs
Fig. 1.
Position of the 11 SNPs in SIRT1, and pair-wise linkage disequilibrium coefficients (r 2) among 11 SNPs in the present Japanese population
Table 2.
Association between SNPs in SIRT2 and diabetic nephropathy
| Allele frequencies (nephropathy case−control) | |||||||
|---|---|---|---|---|---|---|---|
| Proteinuria | ESRD | Combined | |||||
| Study 1 | Study 2 | P | OR (95% CI) | Study 3 | P | OR (95% CI) | |
| SNP | |||||||
| rs1001413a G>A | 0.381/0.359 | 0.353/0.361 | 0.594 | 1.03 (0.91–1.17) | 0.342/0.389 | 0.837 | 0.99 (0.88–1.11) |
| rs892034a C>T | 0.193/0.166 | 0.180/0.169 | 0.099 | 1.14 (0.98–1.34) | 0.164/0.194 | 0.352 | 1.07 (0.93–1.23) |
| rs2015a A>C | 0.398/0.400 | 0.389/0.406 | 0.550 | 0.96 (0.85–1.09) | 0.384/0.391 | 0.522 | 0.96 (0.86–1.08) |
| rs2241703a G>A | 0.235/0.226 | 0.222/0.215 | 0.534 | 1.05 (0.91–1.21) | 0.219/0.208 | 0.453 | 1.05 (0.92–1.20) |
| rs2082435a C>G | 0.247/0.262 | 0.260/0.270 | 0.365 | 0.94 (0.82–1.08) | 0.256/0.231 | 0.678 | 0.97 (0.86–1.10) |
| rs11575003a T>C | 0.118/0.115 | 0.126/0.132 | 0.886 | 0.99 (0.82–1.18) | 0.132/0.119 | 0.887 | 1.01 (0.86–1.19) |
| rs2053071a G>C | 0.377/0.398 | 0.384/0.396 | 0.260 | 0.93 (0.82–1.05) | 0.426/0.402 | 0.506 | 0.96 (0.86–1.08) |
| Haplotype | |||||||
| Block 1 | |||||||
| CCGG | 0.243/0.258 | 0.245/0.268 | 0.136 | 0.90 (0.79–1.03) | 0.254/0.226 | 0.366 | 0.95 (0.84–1.07) |
| CAAC | 0.231/0.222 | 0.227/0.216 | 0.438 | 1.06 (0.92–1.22) | 0.218/0.204 | 0.347 | 1.06 (0.94–1.21) |
| CAGC | 0.179/0.212 | 0.207/0.206 | 0.186 | 0.91 (0.78–1.05) | 0.232/0.205 | 0.477 | 0.95 (0.84–1.09) |
| TAGC | 0.191/0.165 | 0.191/0.169 | 0.037 | 1.18 (1.01–1.37) | 0.164/0.197 | 0.196 | 1.10 (0.95–1.26) |
| CCGC | 0.154/0.142 | 0.127/0.140 | 0.993 | 0.999 (0.84–1.18) | 0.130/0.164 | 0.497 | 0.95 (0.81–1.04) |
| Block 2 | |||||||
| TG | 0.620/0.597 | 0.614/0.599 | 0.191 | 1.08 (0.96–1.22) | 0.568/0.585 | 0.346 | 1.05 (0.95–1.17) |
| TC | 0.262/0.288 | 0.256/0.270 | 0.142 | 0.91 (0.79–1.03) | 0.300/0.295 | 0.121 | 0.91 (0.81–1.02) |
| CC | 0.114/0.110 | 0.126/0.127 | 0.850 | 1.02 (0.85–1.22) | 0.127/0.109 | 0.586 | 1.05 (0.89–1.23) |
Block 1; rs892034, rs2015, rs2241703, rs2082435
Block 2; rs11575003, rs2053071
aTag SNPs
Table 3.
Association between SNPs in SIRT3 and diabetic nephropathy
| Allele frequencies (nephropathy case−control) | |||||||
|---|---|---|---|---|---|---|---|
| Proteinuria | ESRD | Combined | |||||
| Study 1 | Study 2 | P | OR (95% CI) | Study 3 | P | OR (95% CI) | |
| SNP | |||||||
| rs11246002a G>A | 0.137/0.123 | 0.152/0.137 | 0.169 | 1.13 (0.95–1.34) | 0.122/0.110 | 0.138 | 1.13 (0.96–1.32) |
| rs2293168 G>A | 0.356/0.362 | 0.385/0.402 | 0.440 | 0.95 (0.84–1.08) | 0.400/0.372 | 0.776 | 0.98 (0.88–1.10) |
| rs3216 C>G | 0.172/0.168 | 0.160/0.155 | 0.742 | 1.03 (0.87–1.21) | 0.152/0.192 | 0.655 | 0.97 (0.84–1.12) |
| rs10081a A>G | 0.507/0.515 | 0.464/0.463 | 0.805 | 1.02 (0.90–1.15) | 0.460/0.514 | 0.338 | 0.95 (0.85–1.06) |
| rs511744a C>T | 0.488/0.482 | 0.469/0.485 | 0.778 | 0.98 (0.87–1.11) | 0.491/0.487 | 0.853 | 0.99 (0.89–1.10) |
| rs6598074 T>C | 0.164/0.161 | 0.126/0.135 | 0.797 | 0.98 (0.82–1.16) | 0.154/0.144 | 0.963 | 0.996 (0.86–1.16) |
| rs4758633a G>A | 0.347/0.355 | 0.288/0.294 | 0.599 | 0.97 (0.85–1.10) | 0.319/0.349 | 0.360 | 0.94 (0.84–1.06) |
| rs11246007a C>T | 0.143/0.155 | 0.149/0.152 | 0.471 | 0.94 (0.79–1.11) | 0.142/0.143 | 0.512 | 0.95 (0.82–1.11) |
| rs3782117a A>G | 0.168/0.171 | 0.160/0.153 | 0.843 | 1.02 (0.86–1.20) | 0.152/0.193 | 0.544 | 0.96 (0.83–1.11) |
| rs3782116a G>A | 0.307/0.294 | 0.278/0.272 | 0.507 | 1.05 (0.92–1.19) | 0.292/0.268 | 0.333 | 1.06 (0.94–1.20) |
| rs3782115a C>T | 0.283/0.283 | 0.265/0.257 | 0.785 | 1.02 (0.89–1.17) | 0.263/0.241 | 0.964 | 1.04 (0.92–1.17) |
| rs1023430a A>G | 0.291/0.302 | 0.325/0.307 | 0.849 | 1.01 (0.89–1.15) | 0.285/0.275 | 0.743 | 1.02 (0.91–1.15) |
| rs536715a G>A | 0.367/0.367 | 0.395/0.406 | 0.736 | 0.98 (0.86–1.11) | 0.404/0.389 | 0.939 | 0.996 (0.89–1.11) |
| rs3829998a G>A | 0.167/0.167 | 0.124/0.139 | 0.529 | 0.95 (0.80–1.12) | 0.160/0.153 | 0.674 | 0.97 (0.83–1.13) |
| Haplotype | |||||||
| Block 1 | |||||||
| GACT | 0.354/0.362 | 0.378/0.398 | 0.342 | 0.94 (0.83–1.06) | 0.403/0.377 | 0.635 | 0.97 (0.87–1.09) |
| GGCC | 0.335/0.346 | 0.310/0.309 | 0.700 | 0.98 (0.86–1.11) | 0.317/0.321 | 0.688 | 0.97 (0.87–1.09) |
| GGGC | 0.172/0.168 | 0.159/0.154 | 0.688 | 1.03 (0.88–1.21) | 0.151/0.192 | 0.678 | 0.97 (0.84–1.12) |
| AGCT | 0.138/0.123 | 0.151/0.137 | 0.171 | 1.27 (0.95–1.34) | 0.124/0.110 | 0.127 | 1.13 (0.97–1.11) |
| Block 2 | |||||||
| TGGA | 0.519/0.511 | 0.560/0.556 | 0.710 | 1.02 (0.91–1.15) | 0.550/0.521 | 0.462 | 1.04 (0.94–1.16) |
| TAGG | 0.171/0.169 | 0.158/0.153 | 0.765 | 1.02 (0.87–1.20) | 0.151/0.192 | 0.622 | 0.96 (0.84–1.11) |
| TGAA | 0.143/0.155 | 0.150/0.152 | 0.49 | 0.94 (0.80–1.11) | 0.142/0.142 | 0.540 | 0.95 (0.82–1.11) |
| CAGA | 0.167/0.164 | 0.131/0.136 | 0.952 | 0.99 (0.84–1.17) | 0.157/0.146 | 0.868 | 0.95 (0.82–1.11) |
| Block 3 | |||||||
| AAG | 0.364/0.363 | 0.383/0.402 | 0.547 | 0.96 (0.85–1.09) | 0.403/0.384 | 0.779 | 0.98 (0.88–1.10) |
| GGG | 0.287/0.297 | 0.320/0.303 | 0.801 | 1.02 (0.89–1.16) | 0.281/0.265 | 0.640 | 1.03 (0.92–1.15) |
| AGG | 0.177/0.170 | 0.157/0.152 | 0.618 | 1.04 (0.89–1.22) | 0.154/0.191 | 0.809 | 0.98 (0.85–1.13) |
| AGA | 0.168/0.166 | 0.133/0.140 | 0.856 | 0.98 (0.84–1.16) | 0.158/0.152 | 0.967 | 0.997 (0.86–1.16) |
Block 1; rs11246002, rs2293168, rs3216, rs10081
Block 2; rs6598074, rs4758633, rs11246007, rs3782117
Block 3; rs1023430, rs536715, rs3829998
aTag SNPs
Table 4.
Association between SNPs in SIRT4 and diabetic nephropathy
| Allele frequencies (nephropathy case−control) | |||||||
|---|---|---|---|---|---|---|---|
| Proteinuria | ESRD | Combined | |||||
| Study 1 | Study 2 | P | OR (95% CI) | Study 3 | P | OR (95% CI) | |
| SNP | |||||||
| rs6490288 G>C | 0.068/0.076 | 0.076/0.077 | 0.574 | 0.94 (0.74–1.18) | 0.080/0.066 | 0.880 | 0.98 (0.80–1.21) |
| rs7298516a T>G | 0.009/0.009 | 0.008/0.011 | 0.608 | 0.85 (0.46–1.58) | 0.017/0.016 | 0.714 | 0.91 (0.54–1.53) |
| rs3847968a C>T | 0.187/0.184 | 0.187/0.174 | 0.450 | 0.91 (0.71–1.16) | 0.180/0.173 | 0.806 | 1.03 (0.82–1.28) |
| rs12424555 C>T | 0.059/0.069 | 0.065/0.069 | 0.366 | 0.89 (0.70–1.14) | 0.071/0.046 | 0.912 | 0.99 (0.79–1.23) |
| rs7137625a C>T | 0.057/0.040 | 0.058/0.056 | 0.141 | 1.23 (0.94–1.60) | 0.045/0.063 | 0.435 | 1.10 (0.87–1.40) |
| rs2261612 A>G | 0.473/0.484 | 0.457/0.476 | 0.338 | 0.94 (0.84–1.06) | 0.476/0.459 | 0.532 | 0.97 (0.87–1.08) |
| rs2070873a T>G | 0.469/0.476 | 0.457/0.474 | 0.443 | 0.95 (0.85–1.08) | 0.480/0.468 | 0.600 | 0.97 (0.87–1.08) |
| Haplotype | |||||||
| Block 1 | |||||||
| CCCAT | 0.527/0.518 | 0.546/0.520 | 0.245 | 1.07 (0.95–1.21) | 0.517/0.532 | 0.400 | 1.05 (0.94–1.16) |
| CCCGG | 0.350/0.368 | 0.326/0.348 | 0.154 | 0.91 (0.81–1.03) | 0.360/0.342 | 0.305 | 0.94 (0.84–1.05) |
| TTCGG | 0.058/0.067 | 0.065/0.062 | 0.695 | 0.95 (0.75–1.21) | 0.067/0.052 | 0.932 | 1.01 (0.81–1.26) |
| CCTGG | 0.056/0.039 | 0.056/0.056 | 0.181 | 1.20 (0.92–1.56) | 0.046/0.063 | 0.501 | 1.08 (0.86–1.38) |
Block 1; rs3847968, rs12424555, rs7137625, rs2261612, rs2070873
aTag SNPs
Table 5.
Association between SNPs in SIRT5 and diabetic nephropathy
| Allele frequencies (nephropathy case−control) | |||||||
|---|---|---|---|---|---|---|---|
| Proteinuria | Combined | ||||||
| Study 1 | Study 2 | P | OR (95% CI) | Study 3 | P | OR (95% CI) | |
| SNP | |||||||
| rs9382227a G>T | 0.188/0.196 | 0.218/0.192 | 0.494 | 1.05 (0.91–1.22) | 0.198/0.210 | 0.688 | 1.03 (0.90–1.18) |
| rs2804916a T>C | 0.170/0.166 | 0.157/0.163 | 0.921 | 0.99 (0.84–1.17) | 0.138/0.135 | 0.971 | 0.997 (0.86–1.16) |
| rs2804918a A>G | 0.345/0.357 | 0.352/0.333 | 0.847 | 1.01 (0.89–1.15) | 0.318/0.321 | 0.896 | 1.01 (0.90–1.13) |
| rs9370232a G>C | 0.361/0.370 | 0.358/0.362 | 0.640 | 0.97 (0.86–1.10) | 0.357/0.346 | 0.797 | 0.99 (0.88–1.10) |
| rs4712047a G>A | 0.494/0.477 | 0.448/0.505 | 0.221 | 0.93 (0.82–1.05) | 0.456/0.457 | 0.269 | 0.94 (0.84–1.05) |
| rs3734674 G>A | 0.158/0.171 | 0.191/0.149 | 0.252 | 1.10 (0.93–1.29) | 0.176/0.188 | 0.416 | 1.06 (0.92–1.23) |
| rs11751539a A>T | 0.309/0.320 | 0.302/0.312 | 0.476 | 0.95 (0.84–1.09) | 0.315/0.276 | 0.955 | 0.997 (0.87–1.12) |
| rs3757261a G>A | 0.155/0.165 | 0.184/0.139 | 0.159 | 1.12 (0.95–1.32) | 0.168/0.174 | 0.252 | 1.09 (0.94–1.26) |
| rs2253217a A>G | 0.063/0.071 | 0.056/0.068 | 0.210 | 0.85 (0.67–1.09) | 0.045/0.061 | 0.111 | 0.83 (0.67–1.04) |
| Haplotype | |||||||
| Block 1 | |||||||
| GT | 0.641/0.637 | 0.629/0.645 | 0.666 | 0.97 (0.86–1.10) | 0.665/0.655 | 0.796 | 0.99 (0.88–1.10) |
| TT | 0.189/0.196 | 0.215/0.192 | 0.519 | 1.05 (0.91–1.22) | 0.198/0.210 | 0.711 | 1.03 (0.90–1.17) |
| GC | 0.171/0.167 | 0.156/0.163 | 0.086 | 0.87 (0.74–1.02) | 0.137/0.135 | 0.949 | 0.995 (0.86–1.15) |
| Block 2 | |||||||
| GAGA | 0.471/0.442 | 0.446/0.478 | 0.904 | 0.99 (0.88–1.12) | 0.468/0.491 | 0.674 | 0.98 (0.88–1.09) |
| GTGA | 0.311/0.320 | 0.313/0.312 | 0.758 | 0.98 (0.86–1.11) | 0.313/0.272 | 0.734 | 1.02 (0.91–1.14) |
| AAAA | 0.154/0.166 | 0.184/0.139 | 0.150 | 1.12 (0.96–1.32) | 0.169/0.174 | 0.239 | 1.09 (0.94–1.26) |
| GAGG | 0.061/0.067 | 0.054/0.061 | 0.353 | 0.89 (0.70–1.14) | 0.042/0.050 | 0.280 | 0.88 (0.70–1.11) |
Block 1; rs9382227, rs2804916
Block 2; rs3734674, rs11751539, rs3757261, rs2253217, rs2841514
aTag SNPs
Table 6.
Association between SNPs in SIRT6 and diabetic nephropathy
| Allele frequencies (nephropathy case−control) | |||||||
|---|---|---|---|---|---|---|---|
| Proteinuria | ESRD | Combined | |||||
| Study 1 | Study 2 | P | OR (95% CI) | Study 3 | P | OR (95% CI) | |
| SNP | |||||||
| rs350852a T>C | 0.313/0.338 | 0.313/0.303 | 0.545 | 0.96 (0.84–1.09) | 0.324/0.348 | 0.367 | 0.95 (0.84–1.06) |
| rs7246235a T>G | 0.185/0.186 | 0.168/0.209 | 0.110 | 0.88 (0.75–1.03) | 0.202/0.164 | 0.447 | 0.95 (0.82–1.09) |
| rs107251a C>T | 0.296/0.315 | 0.305/0.291 | 0.841 | 0.99 (0.87–1.12) | 0.323/0.328 | 0.799 | 0.98 (0.88–1.11) |
| rs350844 G>A | 0.304/0.322 | 0.309/0.291 | 0.936 | 0.99 (0.87–1.13) | 0.336/0.347 | 0.819 | 0.99 (0.88–1.11) |
| Haplotype | |||||||
| Block 1 | |||||||
| TCG | 0.516/0.499 | 0.529/0.500 | 0.122 | 1.10 (0.98–1.24) | 0.517/0.532 | 0.342 | 1.05 (0.95–1.17) |
| TTA | 0.299/0.318 | 0.303/0.291 | 0.776 | 0.98 (0.86–1.12) | 0.360/0.342 | 0.713 | 0.98 (0.87–1.10) |
| GCG | 0.185/0.183 | 0.168/0.209 | 0.100 | 0.88 (0.76–1.02) | 0.067/0.052 | 0.433 | 0.95 (0.83–1.09) |
Block 1; rs7246235, rs107251, rs350844
aTag SNPs
Table 7.
Replication study for the association between SNPs in SIRT1 and diabetic nephropathy
| Allele frequencies (nephropathy case−control) | Proteinuria (study 1, 2, 4) | Proteinuria + ESRD (study 1, 2, 3, 4) | |||
|---|---|---|---|---|---|
| Study 4 | P | OR (95% CI) | P | OR (95% CI) | |
| SNP | |||||
| rs12778366a T>C | 0.089/0.131 | 0.676 | 0.96 (0.81–1.15) | 0.448 | 0.94 (0.80–1.10) |
| rs3740051a A>G | 0.311/0.291 | 0.226 | 1.08 (0.96–1.21) | 0.106 | 1.09 (0.98–1.22) |
| rs2236318a T>A | 0.113/0.116 | 0.350 | 0.92 (0.78–1.09) | 0.257 | 0.91 (0.78–1.07) |
| rs2236319 A>G | 0.360/0.344 | 0.142 | 1.09 (0.97–1.22) | 0.044 | 1.12 (1.00–1.24) |
| rs10823108 G>A | 0.358/0.337 | 0.127 | 1.09 (0.97–1.23) | 0.038 | 1.12 (1.01–1.24) |
| rs10997868a C>A | 0.181/0.175 | 0.490 | 1.05 (0.91–1.21) | 0.456 | 1.05 (0.92–1.20) |
| rs2273773 T>C | 0.364/0.342 | 0.239 | 1.07 (0.95–1.20) | 0.085 | 1.10 (0.99–1.22) |
| rs3818292 A>G | 0.358/0.344 | 0.120 | 1.10 (0.98–1.23) | 0.040 | 1.12 (1.01–1.24) |
| rs3818291 G>A | 0.090/0.132 | 0.696 | 0.97 (0.81–1.15) | 0.412 | 0.94 (0.80–1.10) |
| rs4746720a T>C | 0.371/0.361 | 0.084 | 0.90 (0.81–1.01) | 0.044 | 0.90 (0.81–0.997) |
| rs10823116a A>G | 0.453/0.450 | 0.939 | 0.996 (0.89–1.11) | 0.446 | 1.04 (0.94–1.15) |
| Haplotype | |||||
| TGTGACCGGTG | 0.306/0.297 | 0.240 | 1.07 (0.95–1.21) | 0.098 | 1.09 (0.98–1.22) |
| TATAGCTAGCA | 0.269/0.243 | 0.809 | 0.96 (0.87–1.11) | 0.336 | 0.95 (0.85–1.06) |
| CATAGCTAATA | 0.105/0.129 | 0.741 | 0.97 (0.82–1.15) | 0.496 | 0.95 (0.81–1.10) |
| TAAAGATAGTA | 0.122/0.116 | 0.621 | 0.96 (0.81–1.13) | 0.430 | 0.94 (0.80–1.09) |
| TATAGCTAGCG | 0.095/0.112 | 0.022 | 0.82 (0.69–0.97) | 0.071 | 0.86 (0.74–1.01) |
| TATAGATAGTA | 0.072/0.059 | 0.0091 | 1.34 (1.07–1.66) | 0.0028 | 1.36 (1.11–1.66) |
| TATGACCGGTG | 0.031/0.044 | 0.942 | 1.01 (0.77–1.33) | 0.746 | 1.04 (0.81–1.35) |
aTag SNPs
Discussion
In the present study, we identified that SNPs within SIRT1 were nominally associated with susceptibility to diabetic nephropathy. We also identified one haplotype consisting of the 11 SNPs in SIRT1 had a stronger association with diabetic nephropathy than single SNPs alone.
SIRT1 encodes a member of NAD(+)-dependent histone deacetylase, involved in various nuclear events such as transcription, DNA replication, and DNA repair. Cumulative evidence during the past decade has demonstrated that SIRT1 plays an important role not only in the regulation of aging and longevity, but also in the development and/or progression of age-associated metabolic diseases, such as type 2 diabetes. SIRT1 activation is considered to be a key mediator for favorable effects on lifespan or on metabolic activity in animals under calorie restriction (CR) [21–24]. Recently, Kume et al. [19] reported that mice under 40% CR were protected from the development of glomerular sclerosis in aging mice kidneys through increasing mitochondrial biogenesis caused by sirt1 activation. From these observations, it is suggested that SIRT1 has a pivotal role in the pathogenesis of aging-related metabolic diseases, such as type 2 diabetes or glomerulosclerosis, and a genetic difference in SIRT1 activity among individuals, if it is present, may contribute to conferring susceptibility to these diseases.
In the present study, we identified that SNPs within SIRT1 were nominally associated with diabetic nephropathy, whereas SNPs in other sirtuin families did not show any association with diabetic nephropathy. Combining the present finding with a previous report, SIRT1 may be considered a good new candidate for diabetic nephropathy, although, the role of sirtuin families other than SIRT1 in age-related metabolic diseases has not been well evaluated. The mechanism by which the SIRT1 polymorphism contributes to conferring susceptibility to diabetic nephropathy remains to be elucidated. Since SIRT1 could affect various metabolic activities, the effects of SIRT1 polymorphisms on susceptibility to diabetic nephropathy might be mediated by differences in the metabolic state among individuals, including glycemic control, obesity, blood pressure, etc. We then examined the association between SNPs in SIRT1 and BMI, hemoglobin A1c (HbA1c), fasting plasma glucose, or systolic blood pressure in the present subjects with type 2 diabetes, but we could not observe any association between the SIRT1 SNPs and those quantitative traits (P > 0.05, Supplementary Table 4). In contrast to our present finding, SNPs within the SIRT1, rs7895833 and rs1467568, were shown to be significantly associated with BMI in Dutch populations [25]. We did not examine those SNPs, but the present study includes an SNP in high linkage disequilibrium (LD) to these 2 SNPs (rs10997868; r 2 = 1 and 0.64 to rs1467568 and rs7895833, respectively). Interestingly, there is a dramatic difference in the frequency of the reported protected allele (A allele of rs1467568) between European and Japanese populations (0.25 in the European population vs. 0.841 in the Japanese population, HapMap database, http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=1467568). Since rs10997868 was not associated with either BMI or susceptibility to the disease, ethnic differences may contribute to the discrepancy between the Dutch and Japanese populations, and the contribution of SIRT1 SNPs to BMI, if it is present, is considered very minor in the Japanese population. It has been also reported that SNPs in SIRT1 were associated with energy expenditure in a small number of Finnish healthy nondiabetic offspring of patients with type 2 diabetes [23]. The alleles associated with higher energy expenditure, supposed to be favorable alleles for glucose metabolism, are G for rs3740051, G for rs2236319, and C for rs2273773, respectively; although these alleles increase the risk of diabetic nephropathy in the present Japanese population. From these observations, we speculate that the effects of SIRT1 gene polymorphisms on diabetic nephropathy are independent of these metabolic parameters; however, there are limitations to the present cross-sectional study and further longitudinal prospective studies are required to obtain a precise conclusion. The association between individual SIRT1 SNPs and diabetic nephropathy did not attain statistically significant levels after correction for multiple-testing errors, and a haplotype consisting of 11 SIRT1 SNPs had a stronger association with the disease, suggesting the existence of other true causal variations within this locus. In addition, since nephropathy cases in the present study were at a more advanced stages of diabetic nephropathy, the findings on SNPs and the haplotype within SIRT1 may be applicable mainly to advanced diabetic nephropathy. Therefore, further extensive analyses for this locus, re-sequencing, dense LD mapping, or further confirmation studies are also required to link the SIRT1 locus to the genetic susceptibility of diabetic nephropathy as a whole.
In conclusion, we found that the SNPs and a haplotype within SIRT1 were nominally associated with susceptibility to diabetic nephropathy in four independent Japanese case–control studies. The present data suggest that SIRT1 may be a good candidate for diabetic nephropathy, although the association should be evaluated further in independent studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank the technical staff of the Laboratory for Endocrinology and Metabolism at RIKEN Center for Genomic Medicine for their technical assistances. This work was partly supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan (to S.M.).
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10157-011-0418-0) contains supplementary material, which is available to authorized users.
References
- 1.U.S. Renal Data System, USRDS 2009 Annual Data Report. Atlas of chronic kidney disease and end-stage renal disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD. Accessed 21 July 2010.
- 2.Nakai S, Masakane I, Akiba T, Shigematsu T, Yamagata K, Watanabe Y, et al. Overview of regular dialysis treatment in Japan as of 31 December 2006. Ther Apher Dial. 2008;12:428–456. doi: 10.1111/j.1744-9987.2008.00634.x. [DOI] [PubMed] [Google Scholar]
- 3.Seaquist ER, Goetz FC, Rich S, Barbosa J. Familial clustering of diabetic kidney disease. Evidence for genetic susceptibility to diabetic nephropathy. N Engl J Med. 1989;320:1161–1165. doi: 10.1056/NEJM198905043201801. [DOI] [PubMed] [Google Scholar]
- 4.Quinn M, Angelico MC, Warram JH, Krolewski AS. Familial factors determine the development of diabetic nephropathy in patients with IDDM. Diabetologia. 1996;39:940–945. doi: 10.1007/BF00403913. [DOI] [PubMed] [Google Scholar]
- 5.Krolewski AS, Warram JH, Rand LI, Kahn CR. Epidemiologic approach to the etiology of type 1 diabetes mellitus and its complications. N Engl J Med. 1987;317:1390–1398. doi: 10.1056/NEJM198711263172206. [DOI] [PubMed] [Google Scholar]
- 6.Fava S, Azzopardi J, Hattersley AT, Watkins PJ. Increased prevalence of proteinuria in diabetic sibs of proteinuric type 2 diabetic subjects. Am J Kidney Dis. 2000;35:708–712. doi: 10.1016/S0272-6386(00)70019-9. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka N, Babazono T, Saito S, Sekine A, Tsunoda T, Haneda M, et al. Association of solute carrier family 12 (sodium/chloride) member 3 with diabetic nephropathy, identified by genome-wide analyses of single nucleotide polymorphisms. Diabetes. 2003;52:2848–2853. doi: 10.2337/diabetes.52.11.2848. [DOI] [PubMed] [Google Scholar]
- 8.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, et al. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005;54:1171–1178. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- 9.Kamiyama M, Kobayashi M, Araki S, Iida A, Tsunoda T, Kawai K, et al. Polymorphisms in the 3′ UTR in the neurocalcin delta gene affect mRNA stability, and confer susceptibility to diabetic nephropathy. Hum Genet. 2007;122:397–407. doi: 10.1007/s00439-007-0414-3. [DOI] [PubMed] [Google Scholar]
- 10.Maeda S, Kobayashi M, Araki S, Babazono T, Freedman BI, Bostrom MA, et al. A single nucleotide polymorphism within the acetyl-coenzyme A carboxylase beta gene is associated with proteinuria in patients with type 2 diabetes. PLoS Genet. 2010;6:e1000842. doi: 10.1371/journal.pgen.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leak TS, Perlegas PS, Smith SG, Keene KL, Hicks PJ, Langefeld CD, et al. Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet. 2009;73:152–159. doi: 10.1111/j.1469-1809.2008.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pezzolesi MG, Katavetin P, Kure M, Poznik GD, Skupien J, Mychaleckyj JC, et al. Confirmation of genetic associations at ELMO1 in the GoKinD collection support its role as a susceptibility gene in diabetic nephropathy. Diabetes. 2009;58:2698–2702. doi: 10.2337/db09-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang SC, Leung VT, Chan LY, Wong SS, Chu DW, Leung JC, et al. The acetyl-coenzyme A carboxylase beta (ACACB) gene is associated with nephropathy in Chinese patients with type 2 diabetes. Nephrol Dial Transplant. 2010;25(12):3931–3934. doi: 10.1093/ndt/gfq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, et al. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeda S, Araki SI, Babazono T, Toyoda M, Umezono T, Kawai K, et al. Replication study for the association between 4 loci identified by a genome-wide association study on European American subjects with type 1 diabetes and susceptibility to diabetic nephropathy in Japanese subjects with type 2 diabetes. Diabetes. 2010;59(8):2075–2079. doi: 10.2337/db10-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kume S, Uzu T, Kashiwagi A, Koya D. SIRT1, a calorie restriction mimetic, in a new therapeutic approach for type 2 diabetes mellitus and diabetic vascular complications. Endocr Metab Immune Disord Drug Targets. 2010;10:16–24. doi: 10.2174/187153010790827957. [DOI] [PubMed] [Google Scholar]
- 18.Liang F, Kume S, Koya D. SIRT1 and insulin resistance. Nat Rev Endocrinol. 2009;5:367–373. doi: 10.1038/nrendo.2009.101. [DOI] [PubMed] [Google Scholar]
- 19.Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 22.Boily G, Seifert EL, Bevilacqua L, He XH, Sabourin G, Estey C, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 24.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zillikens MC, van Meurs JB, Rivadeneira F, Amin N, Hofman A, Oostra BA, et al. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes. 2009;58:2828–2834. doi: 10.2337/db09-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.