Figure 2.
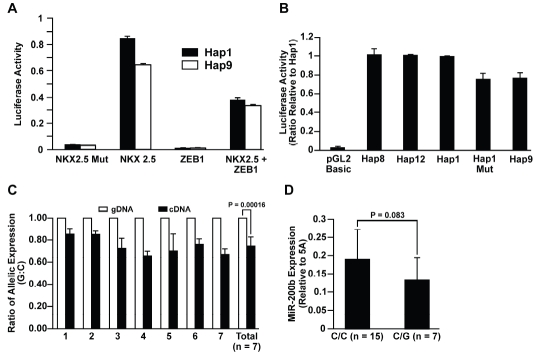
The transcription of miR-200b is affected by rs61768479. Four common haplotypes were identified in the miR-200b-a-429 promoter. The haplotype alleles and frequency are GAGCT (57.5%), GGTCC (18.5%), GGTGC (4.4%), and AGGCC (8.7%) for Hap1, Hap8, Hap9, and Hap12, respectively. The fourth letter bolded is rs61768479. (A) Competition between NKX2.5 and ZEB1 on the promoter activity of miR-200b-a-429 in H1299. NKX2.5 Mut carries two loss-of-function mutants (T178M and K183E) that result in transcriptional inactivation. (B) Luciferase activity for miR-200b-a-429 promoter with different haplotypes in H1299. Hap1 Mut is a mutant form of Hap1 with the NKX2.5 binding site where rs61768479 resides deleted. The luciferase activity for Hap9 and Hap1 Mut was reduced by about 25% as compared to Hap1, Hap8, and Hap12 (P<0.003). No difference was found for luciferase activity between Hap1, Hap8, and Hap12 (P>0.2). (C) rs61768479 dependent allele-specific transcription of miR-200b-a-429. The level of transcription for the G allele is 15–45% lower than that for the C allele (P=0.0002) in seven HBECs heterozygous for rs61768479. The ratio of abundance between G and C alleles in genomic DNA isolated from the same HBEC sample was set as 100%. (D) Expression of mature miR-200b in 7 C/Gs is 25% lower than in 15 homozygotes (P=0.083). Error bars are the standard deviations.