Abstract
The study was aimed at comparing PCR methods of direct detection from biopsy using the boiling method and one other method with two known gold standards (histology and CLO test) for the diagnosis of H. pylori in Nigeria. A total of 168 biopsies (three from antrum and one from corpus each) were taken from 42 patients presenting with various gastroduodenal symptons after informed consent was obtained from them.The biopsies were analysed using the CLO test kit and histology, while the boiling method as described by Holmes and Quigley (1981) was used to obtain DNA and then PCR using the 16S rRNA gene, glmM gene and cagA gene. With CLO test 15/42 (35.71%) were positive, histology 13/42 (30.95%) were positive, 16S rRNA 22/42 (52.38%) were positive, glmM 19/42 (45.24%) were positive, cagA 19/42 (45.24%) were positive. The sensitivity and specificity of the PCR tests with CLO as the gold standard showed that the tests were 100% sensitive and varied between 74.1% to 84.1% in specificity. The PPV and NPV showed that the NPV was almost 100%, while the PPV was between 68.2% and 75%. Using the histology as the gold standard, the sensitivity was almost 100% while the specificity, the PPV were reduced in comparison to the CLO test. The PCR test using the glmM gene appears to be the most reliable test for diagnosis of H. pylori in Nigeria most especially where culture is difficult due to the power outages.
Keywords: PCR, biopsies, CLO test, histology
Introduction
Helicobacter pylori is the causative agent of gastritis, peptic ulcer disease, MALT lymphoma and is a risk factor in the devlopment of gastric cancer [1]. There are several methods for the diagnosis of Helicobacter pylori and can be classed into two broad categories, namely invasive methods that require endoscopy or the minimally or non-invasive methods that do not require endoscopy.
The endoscopy methods include culture, CLO test, PCR, fluorescence in situ hybridization (FISH), direct gram stain, histology, while the non-invasive methods include serology, urea breath test (UBT), Helicobacter pylori stool antigen test(HpSA).
Cutlure is the gold standard for the diagnosis of any microorganism and so where culture cannot be possible histology and CLO test kit have been used as gold standard for the diagnosis of H. pylori in Nigeria. UBT is however the known gold standard for the non-invasive detection of H. pylori.
In Nigeria, currently both gold standards for the non-invasive and invasive detection of H. pylori are fraught with several problems such the method is either difficult for isolation of H. pylori due to frequent power outages in case of culture or very expensive and not generally affordable or available in case of UBT.
This is also in addition to the fact that culture could take several days to obtain a result and in developing countries of which Nigeria is one it is even difficult to obtain a result.
Several assays based on the use of PCR have been developed to detect the presence of H. pylori DNA by using several gene targets directly from the biopsies [2-4]. The targets of these PCR methods include urease A (ureA) gene [5], cag A gene [3], phosphosamine mutase (glmM) gene [4], 16S rRNA gene [3] to mention a few.
The present study was conducted using PCR targeted as some specific genes for H. pylori using the 16S rRNA gene, glmM gene and cagA gene. These method can be standardized to diagnose different agents and part of the standardization has to do with the method of DNA extraction. In addition to the fact that with such methods, all experiments are not lost as in case of power outages and can be easily repeated and not so expensive as the UBT.
The aim of the study was to compare the PCR methods of H. pylori diagnosis with the DNA extraction by Marais et al and the boiling method and compare the results with those of CLO test and histology.
Materials and methods
A total of 126 biopsies from 42 patients (made up of three biopsies each per patients from antrum and one from corpus) were analysed using the following methods: One antrum was taken for histology and another for CLO test while the last antrum sample was taken for culture and PCR.
Histopathologic examination
The gastric biopsies were fixed in 10% buffered formalin for at least 24 h and then embedded in paraffin. In each case, three sections of the tissues were cut at 0.3 micron, de-paraffinized and hydrated in descending grades of alcohol. One section was stained with routine haematoxylin and eosin (H & E) stain following standard procedure [6], the second section was stained with modified Giemsa stain [6] to demonstrate the presence of H. pylori. The organisms were identified as curved rods on the luminal surface of the gastric epithelial cells. The third section was stained with alcian blue/ Periodic Acid Schiff's stain to demonstrate presence of intestinal metaplasia. For classification and grading of gastritis the updated Sydney system [6] was applied. H & E stained slides were used for analyzing histopathological alterations like atrophy, dysplasia, or neoplastic changes as well as erosions and ulcer lesions [6].
DNA extraction
DNA extraction was carried out directly from the biopsies using the boiling method as described by Holmes and Quigley [7]. Briefly, 100μl of the biopsy was added into 200μl of sterile water and vortexed. The samples were then boiled in a dry bath at 100°C for 10 minutes. This was followed by vortexing and centrifugation at 12,000rpm for 5 minutes. The supernatant containing the DNA were transferred to another tube and stored at -20°C. The concentration and purity of the extracted DNA was estimated using a Nanodrop spectrophotometer.
DNA extraction from biopsies was by the method of Marais et al. [8]. Briefly, the biopsy samples were ground and centrifuged for 5 min at 10 000×g. The pellet was resuspended in 300 μL extraction buffer (20 mmol/L Tris-HCl, pH 8.0; 0.5% Tween 20) and proteinase K (0.5 mg/mL final concentration).
The mixture was incubated at 56 for one hour after which the enzyme was inactivated by boiling for 10 mins.
PCR amplification of the 16s rRNA gene
PCR amplification of this gene was carried out using the primer set HEL-F (AAC GAT GAA GCT TCT AGC TTG CTA) and HEL-R (GTG CTT ATT CST NAG ATA CCG TCA T). The 25μl reaction mixture consisted of x1 PCR buffer, 1.5mM Magnesium Chloride, 200μM of each dNTP, 20pmol of each primer and 1U Taq DNA polymerase (Promega).
Amplification was carried out in an Eppendorf Mastercycler gradient using the following cycling parameters. An initial denaturation at 94°C for 5 minutes and 35 cycles of 94°C for 30 seconds, 56°C for 1 minute and 72°C for 1 minute. This was followed by a final extension of 72°C for 10 minutes.
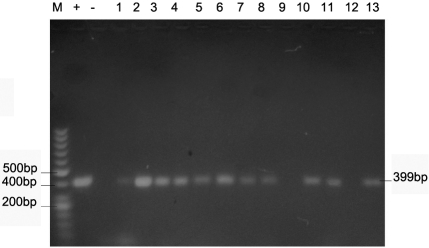
The PCR products were separated on a 2% Aga-rose gel and 50bp ladder was used as DNA molecular weight standard. PCR product size 399 bp.
PCR amplification using the cagA primer
PCR amplification using this primer was carried out using the primer set cagA-F (CCA TGA ATT TTT GAT CCG TTC GG) and cagA-R (GAT AAC AGG CAA GCT TTT GAG GGA). The 25μl reaction mixture consisted of x1 PCR buffer, 1.5mM Magnesium Chloride, 200μM of each dNTP, 20pmol of each primer and 1U Taq DNA polymerase (Promega).
Amplification was carried out in an Eppendorf Mastercycler gradient using the following cycling parameters. An initial denaturation at 94°C for 5 minutes and 40 cycles of 94°C for 1 minute, 54°C for 1 minute and 72°C for 1 minute. This was followed by a final extension of 72°C for 5 minutes.
The PCR products were separated on a 1.5% Agarose gel and 50bp ladder was used as DNA molecular weight standard. PCR product size 349 bp.
PCR amplification using the glmM primer
PCR amplification using this primer was carried out using the primer set glmM-F (GGA TAA GCT TTT AGG GGT GTT AGG GG) and glmM-R (GCT TAC TTT CTA ACA CTA ACG CGC). The 25μl reaction mixture consisted of x1 PCR buffer, 1.5mM Magnesium Chloride, 200μM of each dNTP, 20pmol of each primer and 1U Taq DNA polymerase (Promega).
Amplification was carried out in an Eppendorf Mastercycler gradient using the following cycling parameters. An initial denaturation at 94°C for 5 minutes and 35 cycles of 94°C for 1 minute, 56°C for 1 minute and 72°C for 2 minutes. This was followed by a final extension of 72°C for 5 minutes.
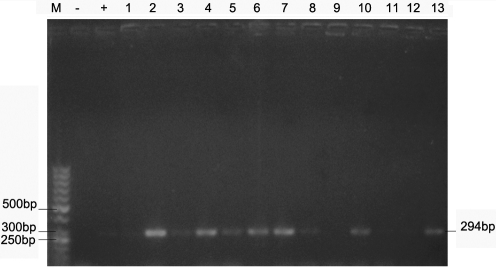
The PCR products were separated on a 1.5% Agarose gel and 50bp ladder was used as DNA molecular weight standard. PCR product size 294 bp.
Results
Endoscopic findings
Out of 42 patients, the endoscopic findings showed that 21/42 (50%) has gastritis, out of which 8/21 (38.1%) were positive for H. pylori in all tests. Endoscopic findings from 42 patients showed that 7/42 (16.67%) were normal findings out of which 3/7 (42.85%) were positive in all three PCR genes, two of which were positive corpus only. Eight (19.05%) of the patients had gastroduodenitis from their endoscopic findings out of which 4/8 (50%) were positive in all three PCR genes and some positive for all the tests, while four (9.52%) had gastric ulcer (GU) from the endoscopic findings, from these 50% were positive in all the tests (Table 1).
Table 1.
Shows the clinical findings and molecular findings of patients presenting with various gastroduodenal symptons in University College Hospital, Ibadan
| Code no | sex | Age | clinical findings/endoscopic findings | Site A & C | CLO (Antrum only) | Histology (antrum only) | 16S rRNA | glmM | cagA |
|---|---|---|---|---|---|---|---|---|---|
| IB 110 | M | 32 | Dyspepsia/normal | A & C | - | - | - | - | - |
| IB 111 | F | 39 | Dyspepsia/gastritis | A & C | + | + | + | + | + |
| IB 112 | M | 33 | Abdominal pain/GORD | A & C | + | + | + | + | + |
| IB 113 | M | 65 | Abdominal pain/ca Stomach | A & C | - | - | C+ | C+ | C+ |
| IB 114 | F | 53 | Acid peptic disease (APD)/gastritis | A & C | - | - | - | - | - |
| IB 115 | M | 26 | Dyspepsia/GU | A & C | - | - | - | - | - |
| IB 116 | F | 44 | Dyspepsia/gastritis | A & C | + | + | A+ | A+ | - |
| IB 117 | M | 73 | Upper GI bleeding/normal | A & C | - | - | C+ | C+ | C+ |
| IB 118 | F | 59 | Dyspepsia/gastritis | A & C | - | - | - | - | - |
| IB 119 | M | 43 | Dyspepsia/gastritis | A & C | - | - | - | - | - |
| IB 120 | M | 24 | Dyspepsia/gastritis | A & C | + | + | + | + | A+ |
| IB 121 | F | 36 | Acid peptic disease/normal | A & C | - | - | - | - | - |
| IB 122 | M | 45 | Acid peptic disease/normal | A & C | - | - | - | - | - |
| IB 123 | F | 42 | Dyspepsia/normal | A & C | - | - | + | + | + |
| IB 124 | F | 57 | Dyspepsia/normal | A & C | - | - | C+ | C+ | C+ |
| IB 125 | M | 49 | Bleeding /normal | A & C | - | - | - | - | - |
| IB 126 | M | 76 | Dyspepsia/gastroduodenitis | A & C | - | - | C+ | + | + |
| IB 127 | F | 50 | Dyspepsia/gastritis | A & C | - | - | - | - | - |
| IB 128 | F | 40 | Dyspepsia/gastritis | A & C | - | - | - | - | - |
| IB 129 | F | 53 | Acid peptic disease/gastroduodenitis | A & C | + | - | + | + | + |
| IB 130 | F | 26 | Acid peptic disease/antral gastritis | A & C | - | - | - | - | - |
| IB 131 | M | 66 | PUD/GU | A & C | + | + | + | + | A+ |
| IB 132 | F | 35 | APD/Gastroduodenitis | A & C | - | - | - | - | - |
| IB 133 | M | 36 | Dyspepsia/gastritis | A & C | + | - | + | + | + |
| IB 134 | M | 54 | Gastric cancer/gastritis/GOO | A & C | + | + | + | A+ | + |
| IB 135 | F | 55 | Upper GI bleeding/erosive gastritis | A & C | - | - | - | - | - |
| IB 136 | F | 50 | Dyspepsia/ GU | A & C | - | - | - | - | - |
| IB 137 | F | 54 | Dyspepsia/gastritis | A & C | + | + | + | + | C+ |
| IB 138 | F | 47 | PUD/GU | A & C | + | + | + | + | + |
| IB 139 | F | 28 | PUD/gastroduodenitis | A & C | + | + | + | + | + |
| IB 140 | F | 57 | Dyspepsia/gastritis | A & C | - | - | - | - | - |
| IB 141 | M | 79 | Chronic DU/gastroduodenitis | A & C | - | - | - | - | - |
| IB 142 | F | 32 | Dyspepsia/gastritis | A & C | + | + | + | + | + |
| IB 143 | M | 47 | Dyspepsia/gastroduodenitis | A & C | + | + | + | + | A+ |
| IB 144 | F | 50 | APD/gastritis | A & C | + | + | + | + | + |
| IB 145 | M | 37 | Gastritis/gastroduodenitis | A & C | - | - | - | - | - |
| IB 146 | F | 60 | PUD/gastritis | A & C | - | - | + | - | - |
| IB 147 | F | 40 | PUD/antral gastritis | A & C | + | + | + | - | + |
| IB 148 | F | 40 | Ca Stomach/gastritis | A & C | - | - | C+ | - | - |
| IB 149 | F | 39 | APD/antral gastritis | A & C | - | - | - | - | - |
| IB 150 | M | 80 | Upper GI bleeding/gastroduodenitis | A & C | - | - | - | - | - |
| IB 151 | F | 33 | APD/corporal gastritis | A & C | - | - | - | - | - |
Keywords: A= antrum, C= corpus, A+= antrum positive only, C+= corpus positive only; PUD= peptic ulcer disease, APD = acid peptic disease, Ca= cancer, GU= gastric ulcer, GOO= gastric outlet obstruction, GORD= gastric oesophageal reflux disease.
The remaining endoscopic findings were GOO, GORD, Ca Stomach accounting for others (4.76%) and all were positive for the three PCR tests and two positive for all the tests. More females than males reported with gastritis (Figure 1).
Figure 1.
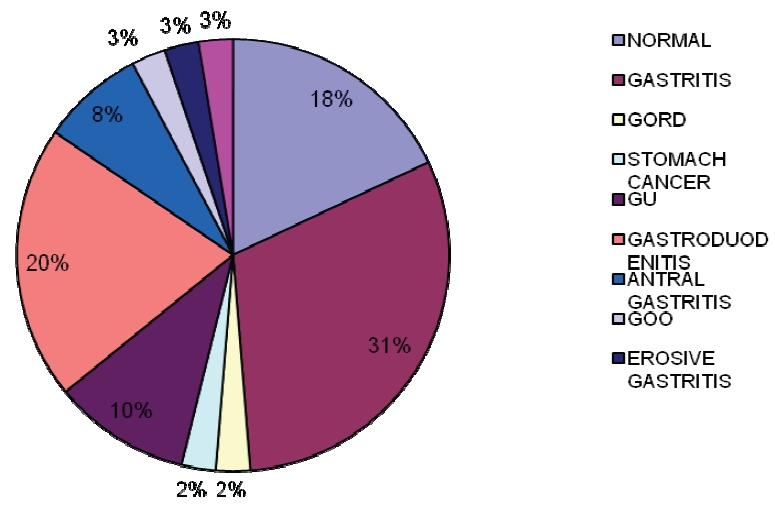
Endoscopic findings of patients presenting with various gastroduodenal symptons.
With the CLO test 15/42 (35.71%), histology 13/42 (30.95%), 16S rRNA 22/42 (52.38%), glmM 19/42 (45.24%), cagA 19/42 (45.24%).
16S rRNA corpus positive were five, glmM gene corpus positive were three. All three genes had corpus positive in three and if it is based on that then the CLO test would give us a total 18/41 (43.9%), while histology would be 16/42 (38.1%).
According to method of DNA extraction by Marais et al. [8], H. pylori DNA was present in 4/42 (9.5%) for cagA and glmM genes while with the 16S rRNA gene, H. pylori was positive in 6/42 (14.3%).
Figures 2-4 show the PCR amplification of H. pylori DNA from biopsy using the cagA, 16S rRNA and glmM genes by the boiling method of extraction.
Figure 2.
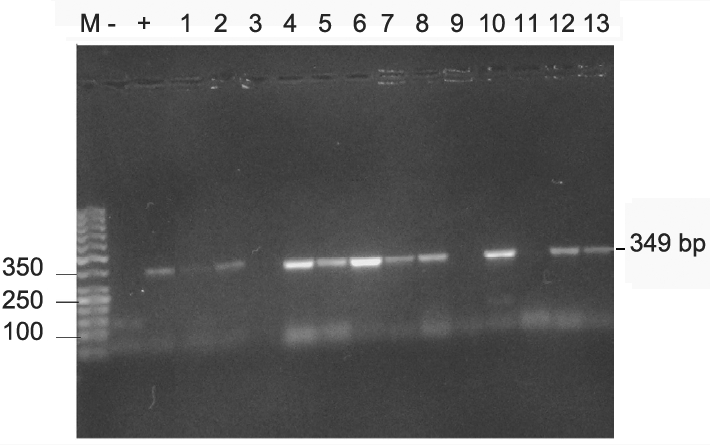
(A) PCR of H. pylori using primers targeted at the cagA gene. Lane M, 50 bp molecular weight marker; Lanes 1 and 6 IB 111 A & C; Lanes 2 and 7, IB 112 A & C; Lane 3 IB 116 A; Lanes 4 & 11, IB 120 A &C, Lanes 5 and 12 IB 123 A & C; Lane 8 IB 113 B, Lane 9 IB 115C, Lane 10 IB 117 B; Lane 13, IB 124C. (B) Lane M, 50 bp molecular weight marker, Lane -, negative control, lane + positive control, lanes 1 & 2 IB 111 A & B, Lanes 3 & 4, IB 112 A & B, Lanes 5 & 6, 123 A & C, Lane 7, IB 120 A, Lane 8, IB 124 C.
Figure 4.
PCR of H. pylori using primers targeted at the glmM gene.
Figure 3.
PCR of H. pylori using primers targeted at the 16S rRNA gene. Lanes M, 50 bp molecular weight marker; Lanes 1 and 6 IB 111 A & C; Lanes 2 and 7, IB 112 A & C; Lane 3 IB 116 A; Lanes 4 & 11, IB 120 A &C, Lanes 5 and 12 IB 123 A & C; Lane 8 IB 113 B, Lane 9 IB 115C, Lane 10 IB 117 C; Lane 13, IB 124C.
Sensitivity, specificity, PPV, NPV
The PCR tests generally showed that the three PCR tests were 100% sensitive and varied in specificity with CLO test as the gold standard while the NPV was almost 100% in the tests (Table 2).
Table 2.
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of PCR
| Value | 16S rRNA gene | glmM gene | cagA gene |
|---|---|---|---|
| Sensitivity | 100% | 100% | 100% |
| Specificity | 74.1% | 81.5% | 81.5% |
| PPV | 68.2% | 75% | 75% |
| NPV | 100% | 95.7% | 95.7% |
However, using histology as the gold standard, the three PCR tests showed the sensitivity to be almost 100% while the specificity, PPV were greatly reduced in comparison to the CLO test (Table 3).
Table 3.
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of PCR specific genes with histology as gold standard.
| Value | 16S rRNA gene | glmM gene | cagA gene |
|---|---|---|---|
| Sensitivity | 100% | 92.9% | 92.9% |
| Specificity | 68% | 78.6% | 78.6% |
| PPV | 59.1% | 68.4% | 68.4% |
| NPV | 100% | 95.7% | 95.7% |
Discussion
The results from the endoscopic findings shows that majority of the patients had gastritis (50%) although most H. pylori cases were from patients with normal findings and gastroduodenitis. A previous report by Smith et al. [9, 10] corroborated this finding that irrespective of their clinical outcome, H. pylori was diagnosed and so also was the cagA gene.
The least sensitive of the methods for diagnosis of H. pylori was histology (30.95%) this could be be due to the type of stain used. In our study the giemsa stain was used for H. pylori diagnosis, but however, from other reports by Ashton-Key et al. [11], diagnosis of H. pylori by histology using different stains affected the sensitivity. From their report, the stain with antibody gave the best detection (66%) as compared with giemsa (55%). In another report by Graham and Graham [12], the choice of stain was also discussed as an important factor in the proper diagnosis of H. pylori.
Most of the results from the CLO test, corroborated with the PCR results obtained and the three cases where CLO test was negative and positive for the other three PCR tests done, was when corpus only was found positive using the PCR tests. The CLO tests were only done on antrum samples and the possibility exists in the fact that probably if the corpus samples were screened for CLO they could have been positive. It was only in one case that there was an inconsistent result between the CLO and the PCR tests. This results differ from a previous report by Smith et al. [13] in which most of the PCR results were negative in comparism to histology and CLO tests. This is because the method of DNA extraction differed from the one used here. The boiling method was used in this procedure while the extraction method as described by Marais et al. [8] was the method employed for DNA extraction. It goes to show also that the method of DNA extraction before PCR was important for the detection of H.pylori DNA in our environment as confirmed from this study (52.38% (16S rRNA gene) and 45.24% (cagA and glmM genes) in comparison to 14.3% for 16S rRNA gene and 9.5% for cagA and glmM genes using the Marais method. In a previous report by Park et al. [14], on the genotyping of H. pylori from biopsy and compared with culture, both methods were approximately equal. Inconsistencies in the two reports was due to low H. pylori density in biopsies. Bjorkholm et al. [15] reported on the use of cagA gene and resistance genes directly from gastric biopsies for H. pylori detection. Another report by Germani et al. [16] showed that the glmM gene was the most specific when compared with culture, histology, 16SrRNA gene and urease test.
In conclusion, the glmM gene using the boiling method of DNA extraction was found to be the most useful method for the direct detection of H. pylori DNA most especially in cases where culture is almost impossible and the UBT is not within the reach of Nigerians due to its affordability.
Acknowledgments
This work was supported by International Centre for Genetic Engineering and Biotechnology (ICGEB) Grant no NIG07-02 to SIS].
References
- 1.Blaser MJ, Perez-Perez GI, Kleanthous H, Cover TL, Peek RM, Chyou PH, Stemmermann GN, Nomura A. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55:2111–15. [PubMed] [Google Scholar]
- 2.Weiss J, Mecca J, da Silva E, Gassner D. Comparison of PCR and other diagnostic techniques for detection of Helicobacter pylori infection in dyspeptic patients. J Clin Microbiol. 1994;32:1663–68. doi: 10.1128/jcm.32.7.1663-1668.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu JJ, Perng CL, Shyu RY, Chen CH, Lou Q, Chong SKF, Lee CH. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J Clin Microbiol. 1999;37:772–774. doi: 10.1128/jcm.37.3.772-774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bickley J, Owen RJ, Fraser AG, Pounder RE. Evaluation of the polymerase chain reaction for detecting the urease C gene of Helicobacter pylori in gastric biopsy sample and dental plaque. J Med Microbiol. 1993;39:338–344. doi: 10.1099/00222615-39-5-338. [DOI] [PubMed] [Google Scholar]
- 5.Clayton CL, Kleanthous H, Coates PJ, Morgan DD, Tabaqchali S. Sensitive detection of Helicobacter pylori by using polymerase chain reaction. J Clin Microbiol. 1992;30:192–200. doi: 10.1128/jcm.30.1.192-200.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis-The updated Sydney System. Am J Surg Path. 1996;20:1161–81. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Holmes DS, Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981;114:193. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- 8.Marais A, Monteiro L, Occhialini M, Pina M, Lamoliatte H, Megraud F. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut. 1999;44:463–467. doi: 10.1136/gut.44.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stella I Smith, Christian Kirsch, Kola S Oyedeji, Anthony O Arigbabu, Akitoye O Coker, Ekkehard Bayerdoffer, Stephan Miehlke. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes in Nigerian patients with duodenal ulcer disease. J Med Microbiol. 2002;51:851–854. doi: 10.1099/0022-1317-51-10-851. [DOI] [PubMed] [Google Scholar]
- 10.Smith SI, Oyedeji KS, Arigbabu AO, Cantet F, Megraud F, Ojo OO, Uwaifo AO, Otegbayo JA, Ola SO, Coker AO. Comparison of three PCR methods for the detection of Helicobacter pylori DNA and detection of cagA gene in gastric biopsy specimen. World J Gastroenterol. 2004;10:1958–1960. doi: 10.3748/wjg.v10.i13.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashton-Key M, Diss TC, Isaacson PG. Detection of Helicobacter pylori in gastric biopsy and resection specimens. J Clin Pathol. 1996;49:107–111. doi: 10.1136/jcp.49.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham KS, Graham DY. 2nd Edition. Published by Handbooks in Health Care Co: Newtown, Pennsylvania, USA; Contemporary diagnosis and management of H. pylori associated gastrointestinal diseases. [Google Scholar]
- 13.Smith S, Omonigbehin E, Goodluck H, Abdulkareem FB, Onyekwere CA, Agomo C, Ndububa DA, Fowora MA, Otegbayo JA, Contreras M, Haas R, Rieder G. Diagnostic methods for the detection of Helicobacter pylori in Nigeria. Trop Gastroenterology. 2010;31:113–115. [PubMed] [Google Scholar]
- 14.Park C-Y, Kwak M, Gutierrez O, Graham DY, Yamaoka Y. Comparison of genotyping Helicobacter pylori directly from biopsy specimens and genotyping from bacterial cultures. J Clin Microbiol. 2003;41:3336–3338. doi: 10.1128/JCM.41.7.3336-3338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjorkholm B, Befrits R, Jaup B, Engstrand Rapid PCR detection of Helicobacter pylori associated virulence and resistance genes directly from gastric biopsy material. J Clin Microbiol. 36:3689–3690. doi: 10.1128/jcm.36.12.3689-3690.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germani Y, Dauga C, Duval P, Huerre M, Levy M, Pialoux G, Sansonetti P, Grimont PA. Strategy for the detection of Helicobacter species by amplificiation of 16S rRNA genes and identification of H. felis in a human gastric biopsy. Res Microbiol. 1997;148:315–326. doi: 10.1016/S0923-2508(97)81587-2. [DOI] [PubMed] [Google Scholar]