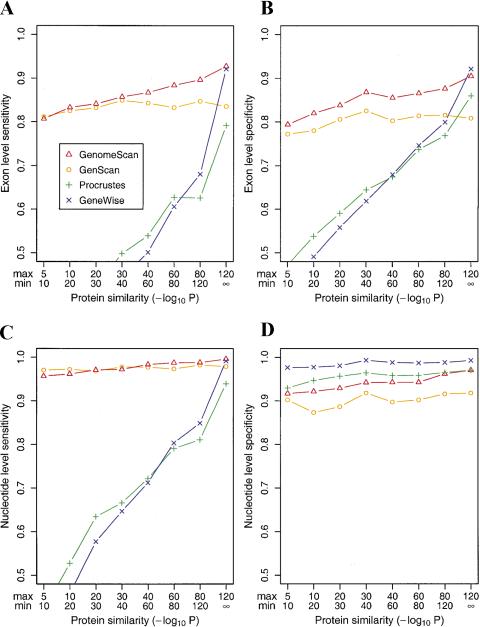
Figure 2.
Exon- and nucleotide-level accuracy of similarity-based gene-prediction programs as a function of protein similarity. (A) Exon-level sensitivity (ESn: percent of exons predicted exactly) and (B) exon-level specificity (ESp: percent of predicted exons exactly correct) were calculated for subsets of the SingleGene dataset and grouped according to the level of BLASTP similarity (in the context of a database search) between the encoded protein and the protein used in the prediction for GenomeScan, Procrustes, and GeneWise as described by Guigó et al. 2000). The definitions of the subsets and number of genes per subset were as follows: 10−5 > P >10−10 (90); 10−10 > P > 10−20 (103); 10−20 > P >10−30 (102); 10−30 > P > 10−40 (97); 10−40 > P >10−60 (114); 10−60 > P > 10−80 (97); 10−80 > P > 10−120 (97); and P < 10−120 (72). For example, 114 of the 175 sequences in the SingleGene dataset had a homolog with BLAST P-value in the range 10−60< P < 10−40. For sequences in this subset, GenomeScan was run using the results of a BLASTX run of the genomic sequence against the top hit in the nonredundant protein database that had sequence similarity in the desired range (10−40 > P > 10−60). GeneWise and Procrustes data, run using the same peptides as input, are from Guigó et al. (2000). (C) Nucleotide-level sensitivity (NSn: percent of coding nucleotides predicted correctly) and (D) nucleotide-level specificity (NSp: percent of predicted coding nucleotides that are correct). Accuracy statistics on the SingleGene dataset as a whole for the ab initio gene-prediction methods GENSCAN, HMMGene 1.1, and GRAIL 3.1, respectively, were as follows: ESn (0.79, 0.75, 0.47); ESp (0.77, 0.68, 0.61); NSn (0.93, 0.86, 0.68): NSp (0.91, 0.74, 0.94).