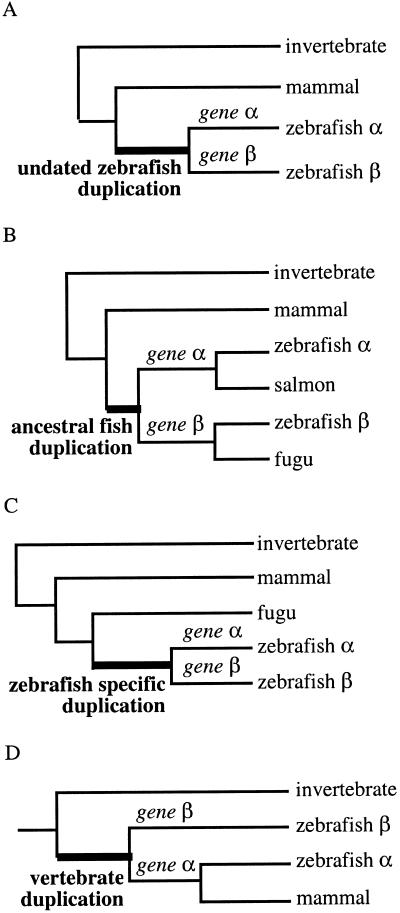
Figure 1.
Determining the origin of duplicated genes by phylogenetic analysis. Branch lengths are arbitrary and do not reflect evolutionary distance. The invertebrate sequence can be replaced by a paralog to root the tree, as described in Methods. Species represented here are arbitrary and depend on sequences available for each gene family. (A) The duplication happened somewhere in the lineage leading to zebrafish. As no other fish sequence is characterized, it may be specific of zebrafish (as in C), or ancestral to fish (as in B). No prediction of the number of genes in other fish is possible. (B) The duplication is shared by several major euteleost fish lineages, proving that the duplication happened in the common ancestor of these fish. The salmon gene should in fact be annotated α, and the fugu gene β. We predict that a salmon β and a fugu α gene should exist, as well as α and β genes in all other euteleost fish, except for secondary losses. (C) The duplication is specific of the zebrafish lineage, and the gene is not duplicated in other major fish lineages. There may be independent duplications in other lineages, but we cannot predict them. (D) The duplication is shared by all vertebrates, even though one of the paralogs was only found in zebrafish. The mammal gene should be annotated α. We predict that a β as well as an α gene should exist in all vertebrates, including mammals, except for secondary losses. Such genes were not used in our analysis.