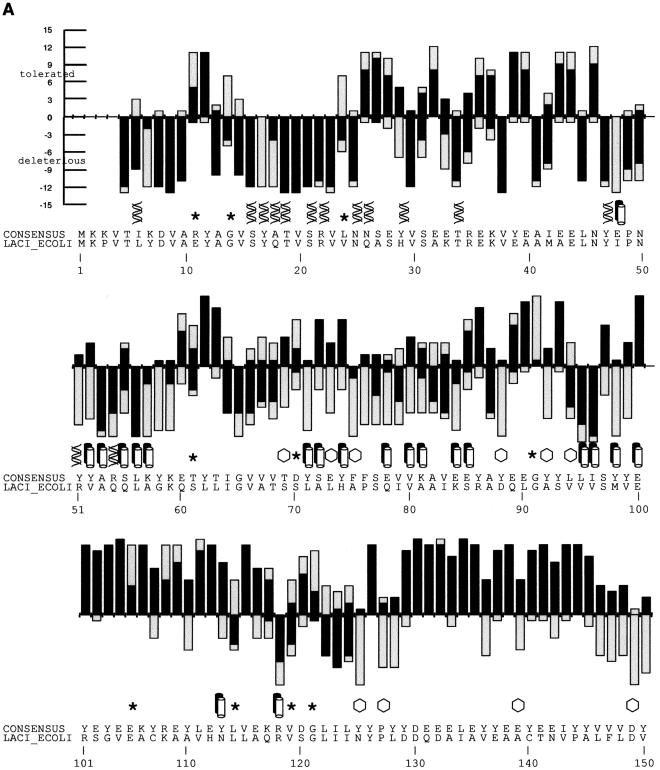
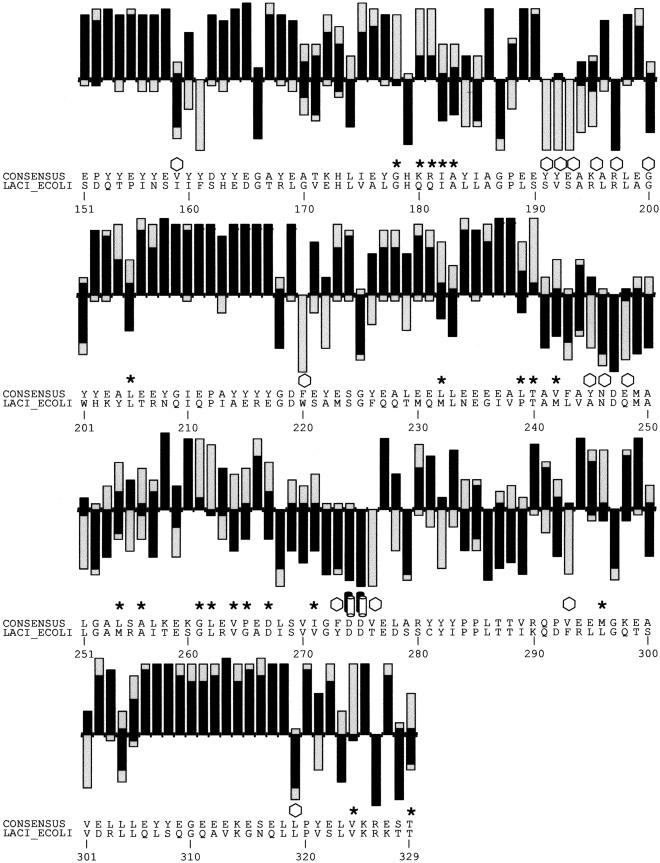
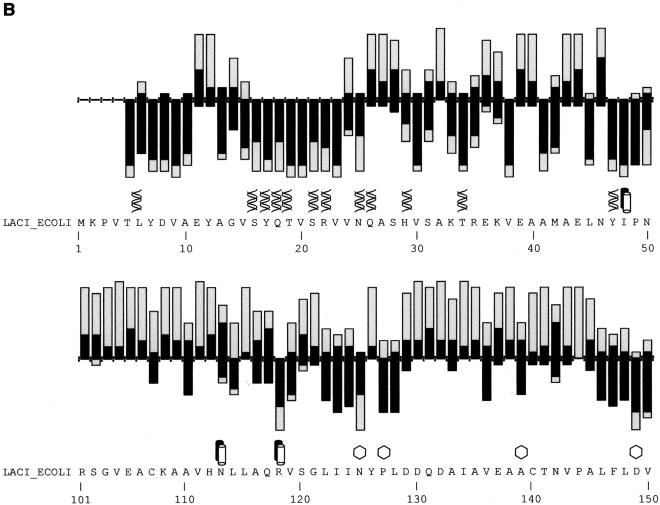
Figure 2.
(A) SIFT predictions for substitutions in LacI. The effects of 12–13 substitutions at each position were assayed (Markiewicz et al. 1994; Suckow et al. 1996). The number of substitutions above the X-axis are those that gave a wild-type phenotype; the number of substitutions below the X-axis gave an affected phenotype. SIFT makes a prediction for every possible substitution, but only substitutions predicted correctly by SIFT are depicted here and are colored in black. Gray bars above the x-axis indicate false positive error; these substitutions were predicted to be deleterious by SIFT, when experimentally they gave wild-type phenotype. Gray bars below the x-axis indicate true negative error; these substitutions were predicted to be neutral, but in fact gave an affected phenotype. Amino acid side chains that have been identified as involved in interactions (Chuprina et al. 1993; Bell and Lewis 2000) are labeled as follows: (double helix) those that interact with DNA, (double cylinders) those participating in the dimer interface. (Hexagons) Positions having six or more substitutions that are unable to respond to the inducer (Markiewicz et al. 1994; Pace et al. 1997). Many of the intolerant positions that were predicted to tolerate substitutions correspond to these query-specific positions. (Asterisks) Positions that can tolerate at least six substitutions, but SIFT predicted more than half of these substitutions as deleterious. The consensus sequence and the original query sequence, LACI_ECOLI, are shown. (B) BLOSUM62 prediction for substitutions in LacI for positions 1–50 and 101–150. BLOSUM62 performs well in the DNA-binding region (residues 1–50) because this region cannot tolerate many substitutions. However, in a region that tolerates substitutions, such as positions 101–150, BLOSUM62 performs poorly, predicting many experimental false positives (large gray bars above the X-axis).