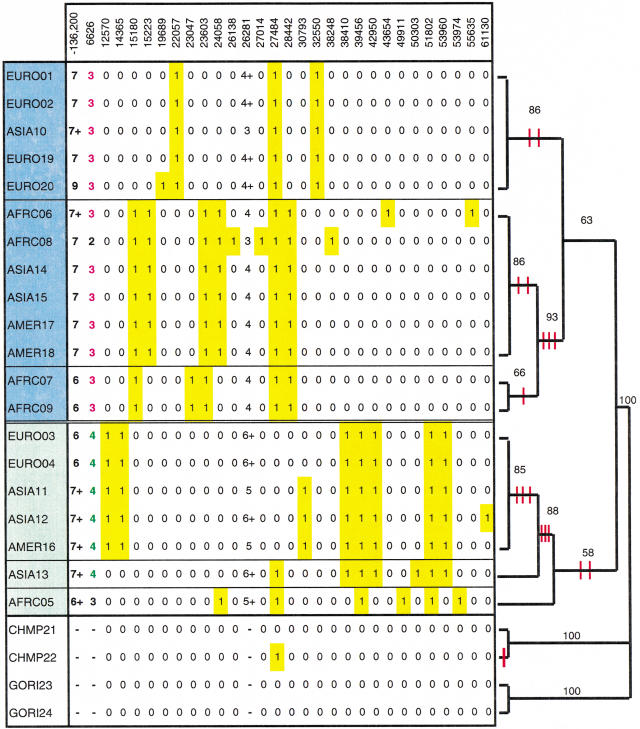
Figure 2.
FMR1 haplotypes grouped according to a branch and bound genealogy based on SNP haplotypes. All derived alleles are highlighted in yellow; microsatellite alleles are listed using the nomenclature in Macpherson et al. (1994) (alleles are given a number correlating to the number of base pairs in the microsatellite repeat tract; a whole number difference indicates a 2-bp increment, whereas a +'sign indicates a 1-bp increment). The sample names shaded in blue and green comprise Clades A and B, respectively. The red hash marks indicate the introduction of single nucleotide changes. The branch and bound algorithm generated five most parsimonious trees, with a tree length of 398, consistency index (CI) of 0.98 (Swofford 1998).