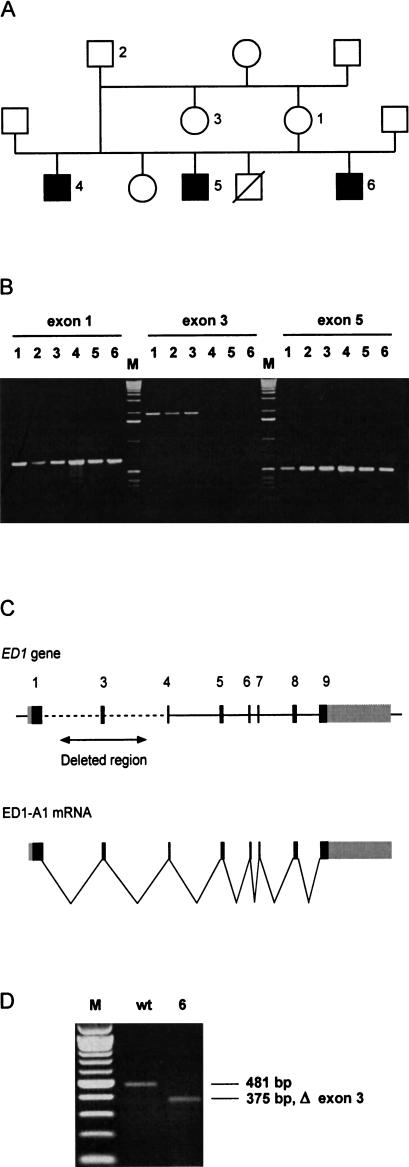
Figure 3.
(A) ED1 cattle family used in this study. In this pedigree, a subset of the animals from a previous study is shown (Drögemüller et al. 2000a). (B) Identification of deletions within the ED1 gene in affected animals. ED1 exons were PCR amplified from animals 1–6 (Fig. 3A). PCR primers flanking exon 3 did not generate PCR products on the DNA of affected animals 4–6, indicating a deletion of this genomic region. (M) 1-kb ladder; (exon 1) 661-bp product, (exon 3) 2006-bp product, (exon 5) 535-bp product with M13-tagged PCR primers). (C) Genomic organization of the mammalian ED1 gene. ED1-A1 and ED1-A2 differ by the presence or absence of 6 nucleotides encoded by exon 8b. (D) RT–PCR amplification of ED1 mRNA. Skin RNA from one affected and one control animal was reverse transcribed and amplified by use of a forward primer located in exon 1 and a reverse primer located in exon 5. The RT–PCR product of the affected animal corresponds to an ED1 cDNA lacking the 106-bp exon 3. The identity of the RT–PCR products and the borders of the deletion were verified by DNA sequencing (M) 100-bp-ladder; (wt) wild type; (6) affected animal No. 6 from Fig. 3A).