Figure 4.
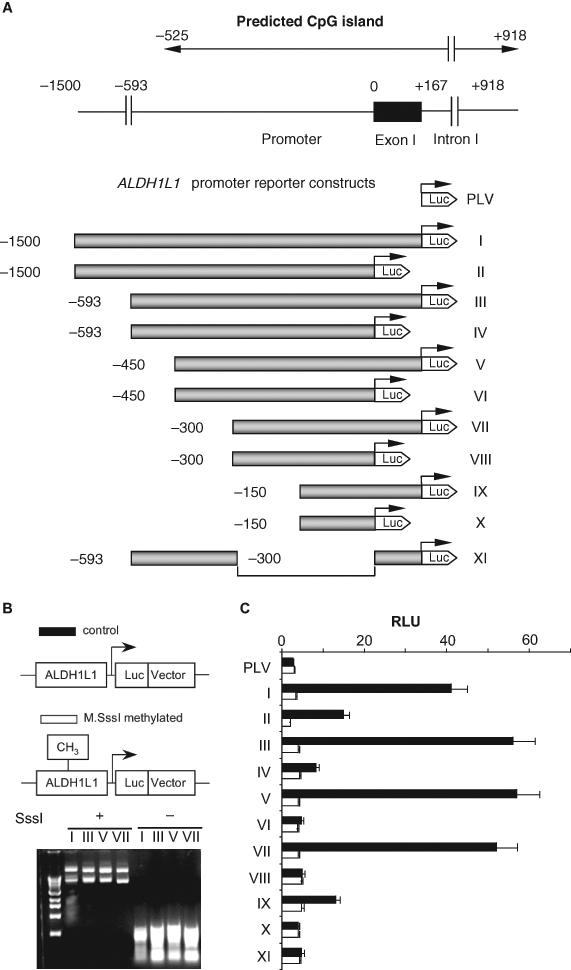
The effect of in vitro methylation on the activity of ALDH1L1 promoter (luciferase assays). (A) Schematic representation of constructs used in a luciferase assay (in each construct, the promoter is shown as a shaded bar, the luciferase gene is shown as a white arrowhead, and the transcription start site is indicated by arrows). Position of the ALDH1L1 CpG island (top diagram) is indicated. (B) Methylation analysis of reporter constructs. Top panel: schematic presentation of methylated and unmethylated plasmids. Bottom panel: agarose gel electrophoresis of methylated (SssI +) and nonmethylated (SssI –) reporter plasmids digested with methylation-sensitive restriction endonucleases, HpaII and HhaI (both enzymes do not cut methylated DNA). (C) Transcriptional activity of ALDH1L1 promoter constructs transiently transfected into A549 cells: closed bars = nonmethylated constructs; open bars = methylated constructs. Luciferase activity (±SD) of 3 independent experiments is shown. PLV = promoterless vector (pGL-basic plasmid).
