Abstract
Using novel monolithic poly(styrene-divinylbenzene) capillary columns with an internal diameter of 0.2 mm, we demonstrate for the first time the feasibility of constructing high-performance liquid chromatography arrays for the detection of mutations by heteroduplex analysis under partially denaturing conditions. In one embodiment, such an array can be used to analyze one sample simultaneously at different temperatures to maximize the detection of mutations in DNA fragments containing multiple discrete melting domains. Alternatively, one may inject different samples onto columns kept at the same effective temperature. Further improvements in throughput can be obtained by means of laser-induced fluorescence detection and the differential labeling of samples with up to four different fluorophores. Major advantages of monolithic capillary high-performance liquid chromatographic arrays over their capillary electrophoretic analogs are the chemical inertness of the poly(styrene-divinylbenzene) stationary phase, the physical robustness of the column bed due to its covalent linkage to the inner surface of the fused silica capillary, and the feasibility to modify the stationary phase thereby allowing the separation of compounds not only on the principle of size exclusion, but also adsorption, distribution, and ion exchange. Analyses times are on the order of a few minutes and turnaround time is extremely short as there is no need for the replenishment of the separation matrix between runs.
High-performance liquid chromatography (HPLC) is unquestionably the most widely used analytical separation technique. The reasons for its popularity are its suitability for separating substances whose chemical properties range from nonpolar to ionic, the availability of a multitude of detection modes such as UV absorbance, refractive index, fluorescence, electrochemical, radioactivity, and mass spectrometric detection, as well as its ready adaptability to accurate quantitative measurements. Over the years, improvements in stationary phase chemistry, such as the synthesis of micropellicular nonporous silica or polymer particles have led to a significant reduction in analysis time from originally hours to a few minutes (Huber 1998). Nevertheless, sample throughput is limited as HPLC does not lend itself readily to the arraying of separation columns as is the case in modern capillary electrophoretic DNA sequencers. Recently, parallel HPLC has been reported primarily as a sample desalting rather than a refined separation procedure prior to mass spectrometric analysis (Bayliss et al. 2000; Feng et al. 2001). In this case, differences in column performance are of little concern. However, for applications of HPLC that depend on reproducibility of chromatographic profiles, such as denaturing HPLC (DHPLC), differences of retention behavior of analytes on different columns become an issue.
Because of its high sensitivity (>96%) and specificity (>99%) in the detection of mutations in DNA fragments as large as 1000 bp, DHPLC has become an increasingly popular tool in the analysis of DNA sequence variation (Xiao and Oefner 2001). Applications of DHPLC include, among others, the discovery of simple sequence polymorphisms (Underhill et al. 1997; Cargill et al. 1999), the mapping of genes (Schriml et al. 2000; Spiegelman et al. 2000a, Hoskins et al. 2001), the mutational analysis of candidate genes (Ophoff et al. 1996), and the targeted screening for induced mutations (Bentley et al. 2000; McCallum et al. 2000), all of which were to benefit from increased throughput.
Recently, it has been shown that monolithic capillary columns synthesized in situ by copolymerization of styrene and divinylbenzene resolve single- and double-stranded DNA fragments with equal to superior separation efficiency as micropellicular poly(styrene-divinylbenzene) columns (Premstaller et al. 2000; Huber et al. 2001). As a consequence of improved peak concentration offered by the capillary format, it became possible to combine DHPLC with argon ion laser-induced fluorescence detection. This allowed a new degree of multiplexing beyond the combined analysis of DNA fragments of different size (Oefner and Underhill 1998). This has been accomplished by labeling amplicons either as a function of the DNA sample used (Xiao et al. 2001), or the genomic region amplified (Xiao and Oefner 2001). To date, up to four different fluorophores commonly used for sequencing and microsatellite genotyping have been combined in a single analysis.
This study demonstrates the feasibility of an HPLC capillary array for the parallel mutational analysis of DNA fragments. Most importantly, individual column temperature adjustments allow compensation of column synthesis-related differences in analyte retention behavior.
RESULTS AND DISCUSSION
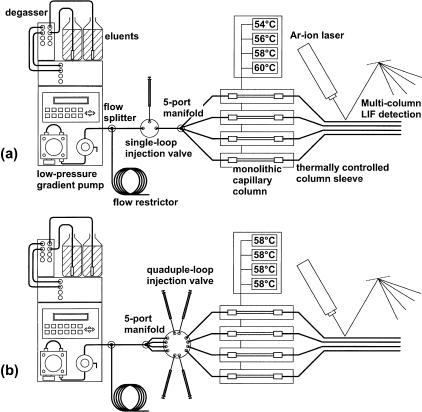
Figure 1 shows a scheme of the two different instrument configurations used. Common to both is the use of a low-pressure gradient-mixing pump operated typically at a flow-rate of 150 μL/min and a back pressure of 16.5 MPa or 2400 psi, an on-line degasser to minimize noise and to prevent fluorescence quenching, and the argon-ion laser-induced fluorescence detector. For the purpose of analyzing a single sample at four different temperatures, an injector with a single internal 1 μL sample loop was placed between a tee and a manifold, both of which were made of stainless steel (Fig. 1a). The tee was used to split the primary flow to create an effective secondary low-flow stream of ∼2.5 μL/min per column. The manifold had a single inlet and four outlets to the columns that were placed inside tubular ovens whose temperatures could be controlled independently from each other. Stainless steel zero dead-volume unions were used to connect the 60 × 0.2 mm i.d. capillary columns to the inlet and outlet fused silica lines with an internal diameter of 25 μm to minimize extra-column volume. The latter should be kept as low as possible to prevent band broadening as a result of longitudinal diffusion of the sample molecules in the mobile phase. To enable monitoring of the fluorescent dye-labeled DNA fragments, a detection window was created by burning off the polyamide layer on the outside of the fused silica capillary.
Figure 1.
Schemes of the two different instrument configurations used to analyze a sample injected once onto four columns (a), and four different samples injected onto four different columns (b). The temperatures of the columns can be regulated individually.
For the loading of four independent samples onto four different columns, the manifold with the single inlet and the four outlets was placed immediately after the tee (Fig. 1b). The four outlet lines were connected to a custom-made electrically actuated injection valve with four internal 1 μL sample loops. Loading was accomplished by injecting 3 μL of sample to ensure complete filling of the loops.
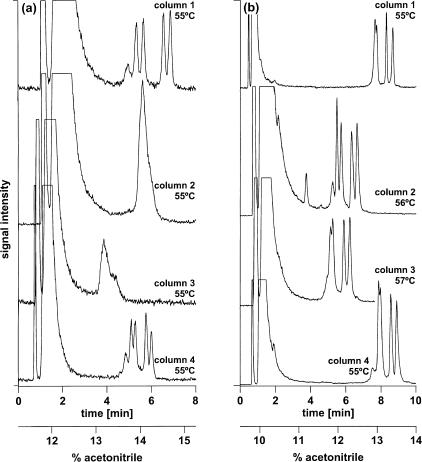
If all columns were identical, they should yield identical chromatograms under identical conditions. However, as can be seen in Figure 2a, the columns yielded different chromatograms. At a uniform column temperature of 55°C, two of the four columns yielded a four-peak profile on injection of a 209-bp amplicon representing two Y-chromosome alleles differing in a single base, an A to G transition, at nucleotide position 168. The first two peaks represent the heteroduplex species formed on mixing, denaturing, and reannealing of two polymorphic chromosomes, whereas the latter two peaks represent the corresponding homoduplexes. The remaining two columns yielded a single peak at 55°C. Using individual column thermostats, the column temperatures of the latter columns could be increased to 56°C and 57°C, respectively, to obtain chromatographic profiles similar to those observed with the former two columns at 55°C (Fig. 2b).
Figure 2.
Adjustment of the chromatographic elution profiles of different columns by means of variation of column temperature. (a) The same sample was injected onto four different columns kept at the same temperature in a commercially available enforced air circulation oven with Peltier cooling. Because of differences in surface area, retention of DNA fragments varies between columns. (b) Variation of the individual column temperatures allows harmonization of chromatographic profiles. Columns, 4 × monolithic PS/DVB, 60 × 0.2 mm i.d., mobile phase, (A) 100 mM TEAA at pH 7.0, (B) 100 mM TEAA at pH 7.0, 25% acetonitrile; linear gradient, (a) 45%–61% B in 8.0 min, (b) 38%–56% B in 10 min; flow rate, 2.0–3.0 μL/min; temperature, (a) 55°C, (b) 55–57°C; detection, LIF, emission monitored at 525 nm; injection volume, 1 μL each; sample, equimolar mixture of two FAM-labeled 209-bp Y chromosome alleles differing in a single base (168 A>G).
The necessity to use different column temperatures to obtain identical chromatographic profiles is the result of differences in retention of nucleic acids between columns. For example, at a column temperature of 50°C, oligodeoxythymidylic acids 12–18 nucleotides in length were retained less, i.e., they eluted at a lower acetonitrile concentration from those columns that required a higher temperature to obtain the separation of all four homo- and heteroduplex species. It was shown recently for nucleic acids labeled with different fluorophores that an ∼0.8% increase in acetonitrile concentration that was required for elution corresponded to an increase in column temperature of 1°C (Xiao and Oefner 2001). Consequently, if a given DNA fragment is eluted from a column at a lower acetonitrile concentration, a higher column temperature is required to achieve the same degree of partial denaturation. This is also evident from Figure 2. At a temperature of 55°C, the amplicon eluted from column #3 at a concentration of ∼13% acetonitrile, whereas the same amplicon eluted from column #2 at 14% acetonitrile (Fig. 2a). Consequently, the temperature of column #3 had to be raised 1°C higher than that of column #2 to obtain a similar separation of the homo- and heteroduplex species.
Retention and, consequently, the concentration of acetonitrile required for eluting a double-stranded DNA fragment from the column is governed mainly by electrostatic interactions between the positively charged amphiphilic triethylammonium ions adsorbed at the surface of the nonpolar stationary phase and the dissociated phosphodiester groups of the sugar-phosphate backbone of DNA. Another important factor is the surface area of the stationary phase, because retention is proportional to the surface area of the adsorbent. Adsorbent surface area increases with decreasing pore size. The column backpressure is an indicator of pore size and increases also with decreasing pore size. Differences in surface area are most likely the result of the instability of divinylbenzene, as differences in the ratio of styrene to divinylbenzene affect the degree of polymerization and, consequently, pore size. Indeed, back-pressure was greater in case of the capillary columns that retained the nucleic acids longer. Therefore, instead of carefully selecting columns with similar porosity and retention characteristics, one may adjust column temperature by means of individual column thermostats until all columns yield very similar chromatographic profiles (Fig. 2b). Subsequent changes in column temperature will have identical effects on peak pattern of all columns. Such harmonization of chromatographic separation by modulation of column temperature is by no means restricted to DHPLC. Rather, it can be applied to any chromatographic separation sensitive to temperature.
The practice of DHPLC has been facilitated by the development of an algorithm that predicts the temperature at which half of the base pairs at a given nucleotide site are no longer hydrogen bonded under chromatographic conditions (Jones et al. 1999). This is considered the optimum temperature to detect a base mismatch at a given site. This algorithm had been optimized for the micropellicular octadecylated poly(styrene/divinylbenzene) chromatographic support that is still the most widely used in DHPLC. However, it was already noticed that column temperature had to be increased 2°C when using the novel monolithic poly(styrene/divinylbenzene) chromatographic support (Huber et al. 2001). Because the monolith is not alkylated, its surface is more polar. Thus, lower concentrations of acetonitrile are required for elution of DNA fragments, which in turn, necessitates a higher column temperature to achieve the same degree of partial denaturation compared to an alkylated separation matrix. To allow the continued use of the melting algorithm in conjunction with capillary arrays, it is necessary to calibrate the columns in the array. For that purpose, the temperature of every column is adjusted with the help of a known mutation until all columns display the same profile that is then compared to profiles of the same mutation generated on a micropellicular support. The temperature that generated the profile most similar to that on the monolith is then coined the effective temperature at which the analysis was carried out.
After columns have been calibrated, reproducible chromatographic separations can be achieved. The mean relative standard deviation of the retention times of the two heteroduplex and the two homoduplex peaks on the four columns over six injections was 1.84%, with a range of 0.83%–2.62% (Table 1). Moreover, reproducibility among the four temperature-controlled sleeves was quite similar ranging from ±1.6% to ±2.1%. Variations in peak profiles constitute a problem when DHPLC is not only used for discovery of DNA variants but also for their genotyping. Usually, reproducibility with conventional HPLC column ovens is so high that in a recent study of 432 BRCA1 and 136 BRCA2 variants, all but two were identified correctly on comparison with peak patterns obtained for known alternations (Gross et al. 2000). However, for all other purposes including the fine mapping of traits, variation of peak patterns is of no concern as long as it does not affect the general ability to detect the variant.
Table 1.
Reproducibility of Retention Times
| Run no.Peak | Column 1 | Column 2 | Column 3 | Column 4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | |
| 1 | 7.50 | 7.74 | 8.53 | 8.88 | 5.33 | 5.69 | 6.31 | 6.37 | 5.15 | 5.45 | 6.11 | 6.35 | 7.45 | 7.64 | 8.15 | 8.58 |
| 2 | 7.60 | 7.68 | 8.47 | 8.81 | 5.63 | 5.83 | 6.43 | 6.73 | 5.37 | 5.57 | 6.28 | 6.63 | 7.78 | 7.91 | 8.48 | 8.87 |
| 3 | 7.67 | 7.80 | 8.31 | 8.67 | 5.49 | 5.74 | 6.33 | 6.65 | 5.15 | 5.28 | 5.91 | 6.25 | 7.88 | 7.98 | 8.57 | 8.91 |
| 4 | 7.61 | 7.68 | 8.11 | 8.63 | 5.60 | 5.82 | 6.38 | 6.67 | 5.41 | 5.62 | 6.18 | 6.42 | 7.82 | 7.92 | 8.53 | 8.91 |
| 5 | 7.67 | 7.73 | 8.09 | 8.57 | 5.58 | 5.83 | 6.41 | 6.75 | 5.27 | 5.48 | 6.12 | 6.43 | 7.87 | 8.00 | 8.27 | 8.58 |
| 6 | 7.58 | 7.58 | 8.01 | 8.45 | 5.61 | 5.94 | 6.55 | 6.81 | 5.15 | 5.43 | 6.10 | 6.36 | 7.81 | 7.96 | 8.46 | 8.90 |
| Mean | 7.61 | 7.70 | 8.25 | 8.67 | 5.54 | 5.81 | 6.40 | 6.66 | 5.25 | 5.47 | 6.12 | 6.41 | 7.77 | 7.90 | 8.41 | 8.79 |
| SD | 0.06 | 0.07 | 0.22 | 0.16 | 0.11 | 0.09 | 0.09 | 0.15 | 0.12 | 0.12 | 0.12 | 0.13 | 0.16 | 0.13 | 0.16 | 0.16 |
| RSD | 0.83 | 0.97 | 2.62 | 1.81 | 2.05 | 1.48 | 1.34 | 2.32 | 2.26 | 2.17 | 1.99 | 1.98 | 2.07 | 1.68 | 1.95 | 1.87 |
Reproducibility of retention times in minutes of the four heteroduplex peaks on a four-column array with four individual resistance ovens at an effective column temperature of 55°C over six runs. Experimental conditions as described in Figure 2b.
The excellent mutation detection sensitivity of DHPLC (approaching almost 100% Xiao and Oefner 2001) is caused in part by the fact that the majority of mutations can be detected over a wide range of temperatures. In a recent study, 32 mutations could be detected over temperature windows spanning on average 8°C, with a range of 4°C –11°C (Jones et al. 1999). However, there have been rare reports of mutations that could be detected only at a unique temperature or over a more narrow temperature window of 2°C –3°C (Wagner et al. 1999; Nickerson et al. 2001). Therefore, in case of multiple melting domains that differ by >5°C, it has been suggested to repeat the analysis in 5°C decrements starting with the highest temperature recommended by the melting algorithm (Jones et al. 1999). This was put to a test in a study of 166 polymorphic DNA fragments corroborating that sensitivity increased from 94% to 99.8% when the samples were analyzed at all recommended temperatures (Spiegelman et al. 2000b). However, because samples can only be injected serially, such repeated analyses at different temperatures reduce overall sample throughput.
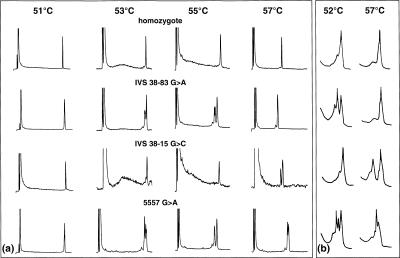
Recently, it has been suggested that the use of an externally controlled temperature gradient >10°C would allow detection of all variants independently of their melting behavior (Gao and Yeung 2000). However, in our hands we found this to apply only to variants with similar melting characteristics as it was the case in the aforementioned study with the melting temperatures of all five investigated single nucleotide polymorphisms (SNPs) ranging from 65°C –69°C. When applied to variants melting only at the extremes of the temperature gradient, the approach failed to detect the polymorphisms known to melt close to the starting point of the gradient. For that reason, we explored an alternative approach injecting 1 μL of sample simultaneously onto four capillary columns that were kept at different temperatures by means of thermally controlled sleeves. Figure 3 depicts the multiple temperature analysis of three different mutations in the coding and flanking noncoding region of exon 39 of the ATM gene as well as that of a homozygous control. The effective temperature used to screen the polymorphic fragments on the monolithic capillary array was increased in 2°C increments from 51°C –57°C. In agreement with the previous serial analysis on the commercially available micropellicular column support (Thorstenson et al. 2001), the G>A transition located 83 base pairs upstream of exon 39 could only be detected at 53°C and 55°C, but not at 57°C. Correspondingly, the G>C transversion 15 base pairs upstream of exon 39 could be detected only at the highest temperature, but not at the lower temperatures. Finally, the G>A transition located within exon 39 could be detected at 53°C, 55°C, and 57°C, respectively. Overall, the results were in agreement with the predicted melting behavior of the variant sites.
Figure 3.
Parallel analysis of amplicons at multiple temperatures. (a) Using a manifold (see Fig. 1a for instrumental details) the sample was split and directed to four monolithic capillary columns kept at four different temperatures. (b) The same samples had been analyzed previously serially on a commercially available micropellicular column support at 52°C and 57°C. Experimental conditions: (a) columns, 4 × monolithic PS/DVB, 60 × 0.2 mm i.d.; mobile phase, (A) 100 mM TEAA at pH 7.0, (B) 100 mM TEAA at pH 7.0, 25% acetonitrile; linear gradient, 45%–61% B in 8.0 min; flow rate, 2.0–3.0 μl/min; temperature, 51°C, 53°C, 55°C, and 57°C, respectively; detection, LIF, emission monitored at 525 nm; injection volume, 1 μL each; (b) column, DNASep, 50 × 4.6 mm i.d.; mobile phase, (A) 100 mM TEAA at pH 7.0, (B) 100 mM TEAA at pH 7.0, 25% acetonitrile; linear gradient, 58%–64% B in 3.0 min at 52°C, and 54%–60% B in 3.0 min at 57°C; flow rate, 900 μL/min; detection, UV, 254 nm; injection volume: 8 μL each. Samples: one homozygous and three different heterozygous samples comprising exon 39 and flanking noncoding regions of ATM.
Alternatively, the array may be used to analyze different samples in parallel using a photodiode array UV absorption detector (Gong and Yeung 1999). Throughput can be increased further by multiplex analysis of amplicons labeled with different fluorophores. There are two different multiplex strategies. First, one can use the fluorophores to distinguish the same amplicon generated from different samples (Xiao et al. 2001). Second, one may label different amplicons generated from the same DNA sample (Xiao and Oefner 2001). In the latter case, one has to make sure that the different amplicons share similar melting characteristics. Moreover, the impact of the different fluorophores on retention has to be considered. It has been shown that the chromatographic profile of a polymorphic FAM-labeled amplicon resembles that of an unlabeled fragment (Xiao et al. 2001). In contrast, the profiles of polymorphic amplicons tagged with the more hydrophobic fluorophores HEX or NED resemble those of unlabeled PCR products analyzed at a 1°C higher column temperature. Finally, the chromatographic pattern of a ROX-labeled amplicon is similar to that of an unlabeled fragment injected at a 2°C higher column temperature. ROX is the most hydrophobic of the four fluorophores and, therefore, it is retained the longest on the nonpolar stationary phase. With increasing retention of the labeled fragments, an increasingly higher concentration of acetonitrile is required for their elution. Because organic solvents reduce the melting temperature of double-stranded DNA, the peak patterns seen with increasing hydrophobicity of the fluorophore resemble those of an unlabeled fragment analyzed at increasing column temperature. Consequently, to ensure maximum mismatch detection sensitivity only fluorophores with similar effects on retention should be chosen. Alternatively, the melting algorithm described earlier may be used to select fragments with higher melting characteristics to compensate for the drop in melting temperature as a function of the fluorophore attached.
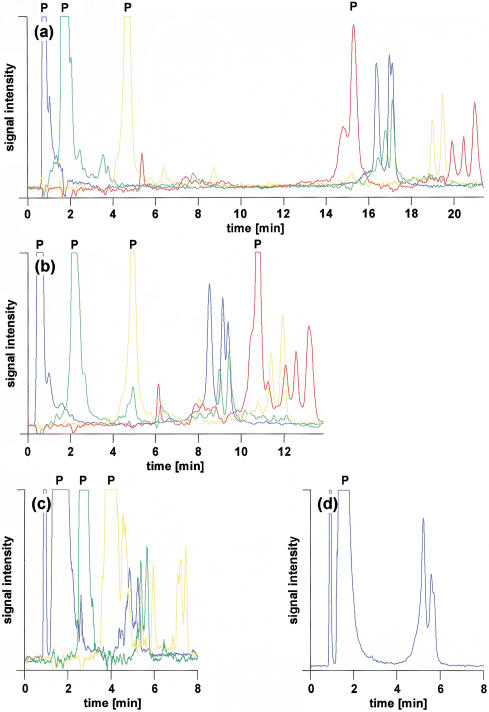
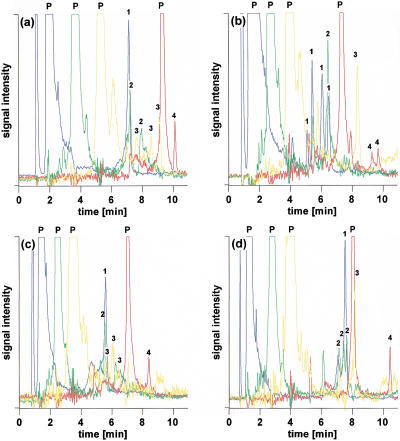
Figure 4a shows the multiplex analysis of four different polymorphic DNA fragments labeled with FAM, HEX, NED, and ROX, respectively. Although primers and amplicons carry one fluorescent dye only, the great hydrophobicity of ROX causes the ROX-labeled primer to be retained approximately as long as amplicons tagged with one of the other three less hydrophobic fluorophores. It is possible to elute all labeled primers first, followed by their corresponding amplicons, using an isocratic flow of 11.75% acetonitrile over 8 min before starting the acetonitrile gradient. However, this results in a total analysis time of ∼25 min, thereby offsetting any gain in throughput accomplished by differential labeling as unlabeled products can be separated in as little as 4 min including regeneration of the column between injections (Xiao and Oefner 2001). Analysis time can be shortened but care has to be taken that the ROX primer does not coelute with one of the amplicons tagged with FAM, HEX, or NED, because it would become impossible to spectrally resolve the signals of the latter from the abundant signal of the ROX-labeled primer. As shown in Figure 4b, optimization of the gradient allowed the successful separation of the ROX-labeled primer from the other components in the sample while cutting the time of analysis in half, hence allowing the loading of the array four times per hour. However, if only FAM, HEX, and NED were used as tags, total analysis time could be reduced to 10 min (Fig. 4c). Consequently, the array could be loaded up to six times per hour, leading to an increase in throughput of 12.5% over the use of four fluorophores and four runs per hour. However, it appears that the close elution of unincorporated NED-labeled primer and FAM-labeled amplicon exerted a detrimental effect on the separation efficiency of the latter. The fronting of the heteroduplex peak suggests competition with the NED-labeled primer for adsorption to the stationary phase. As seen in Figure 4d, the shape of the FAM-labeled hetero- and homoduplex peaks could be restored to that observed in Figure 4, panels a and b, when only the FAM-labeled PCR was injected. For that reason, it appears that five injections per hour are more realistic, which corresponds to a 6% decrease in throughput compared to four runs per hour with four fluorophores. However, this loss in throughput is compensated by the fact that no extensive optimization of the gradient conditions is required to avoid coelution of ROX-labeled primer with other compounds. At present, spectral overlap prevents the use of other available less hydrophobic tags that can be excited with an argon-ion laser, such as TET and TAMRA. Eventually, the use of a multi-color laser or delayed fluorescence detection may allow the inclusion of additional fluorescent dyes, such as Cascade Blue, that also exhibit a favorable retention behavior.
Figure 4.
Impact of gradient profile on the retention and separation efficiency of primers and amplicons labeled with FAM (blue), HEX (green), NED (yellow), and ROX (red), respectively. Column, monolithic PS/DVB, 60 × 0.2 mm i.d.; mobile phase, 100 mM TEAA at pH 7.0; gradient, (a) 11.75% acetonitrile for 8 min, 11.75%–16.0% acetonitrile in 10 min, (b) 11.75%–16.0% acetonitrile in 10 min, (c,d) 12%–16.4% acetonitrile in 7.0 min; flow rate, 2.0–3.0 μL/min; effective temperature, 58°C; detection, LIF, emission monitored at 525, 555, 580, and 605 nm, respectively; injection volume, 1 μL each; sample, MDR1, blue, exon22, 320 G>A heterozygote; green, MDR1, exon 13, 291 T>C heterozygote; yellow, MDR1, exon 26, 325 T>G and 346 T>C double heterozygote; and red, MDR1, exon 26, 212 C>T and 346 T>C double heterozygote.
Figure 5 shows the parallel separation of both homozygous and heterozygous amplicons labeled with FAM, HEX, NED, or ROX, using an array of four capillary columns. However, to ensure reproducible gradient formation, almost all pumps commercially available for running capillary columns use a relatively high primary flow of 100–200 μL/min before splitting the flow with almost 99% of the mobile phase going to waste. Therefore, there is plenty of eluent available to run theoretically up to 96 capillaries. Using existing injector technology, at least another three of the four-column injectors used in the present study can be added, bringing the total number of capillaries up to 16. Using sample pooling that has been shown repeatedly not to affect sensitivity if the number of chromosomes is limited to 10 (McCallum et al. 2000; Wolford et al. 2000) and four fluorescent dyes of similar retention behavior, as many as 3800 amplicons can be processed per hour on a 16-capillary array. Although throughput is still too low to search on a genome-wide scale for induced mutations, it appears sufficient to screen at least hundreds of genes. At the recovery rates of mutant strains reported for Arabidopsis (McCallum et al. 2000) and Drosophila (Bentley et al. 2000) that ranged from ∼2.5 to 5 mutations/kb for every 1000 chromosomes mutagenized by ethyl methanesulfonate, one can expect to detect about 5–10 mutant alleles per hour scanning 600 bp per amplicon.
Figure 5.
Simultaneous analysis of 16 different samples in four capillary columns containing samples that were homozygous or heterozygous for exon 13, 22, or 26 in MDR1. Columns, 4 × monolithic PS/DVB, 60 × 0.2 mm i.d.; mobile phase, (A) 100 mM TEAA at pH 7.0, (B) 100 mM TEAA at pH 7.0, 25% acetonitrile; linear gradient, 45%–70% B in 10.0 min; flow rate, 2.0–3.0 μL/min; temperature, (a,d) 55°C, (b) 56°C, (c) 57°C; detection, LIF, emission monitored at 525 nm for FAM (blue), 555 nm for HEX (green), 580 nm for NED (yellow), and 605 nm for ROX (red); injection volume, 1 μL each; samples, column 1, exon 22, FAM, homozygote, exon 13, HEX, 52 T>G and 291 T>C double heterozygote, exon 26, NED, 212 C>T and 346 T>C double heterozygote, exon 26, ROX, homozygote; column 2, exon 22, FAM, 320 G>A heterozygote, exon 13, HEX, homozygote, exon 26, NED, homozygote, exon 26, ROX, 212 C>T and 346 T>C double heterozygote; column 3, exon 22, FAM, homozygote, exon 13, HEX, homozygote, exon 13, NED, 52 T>G and 291 T>C double heterozygote, exon 26, ROX, homozygote; column 4, exon 22, FAM, homozygote, exon 13, HEX, 52 T>G and 291 T>C double heterozygote, exon 13, NED, homozygote, exon 26, ROX, homozygote.
In conclusion, monolithic poly(styrene/divinylbenzene) capillary columns allow the construction of arrays similar to those reported for capillary electrophoresis. More than 500 injections have been performed on the capillaries used in this study without any notable deterioration in column performance and they will likely prove as durable as conventional micropellicular poly(styrene/divinylbenzene) columns. Laser-induced fluorescence detection enables a further increase in sample throughput by differential labeling of DNA fragments with fluorophores commonly used in DNA sequencing. Differences in retention behavior between columns can be neutralized by adjusting the temperature of each column. Such arrays hold the promise of transforming DHPLC into a true genome scale technology.
METHODS
DNA Sequences and Primers
A 209-bp amplicon containing either an A or a G at nucleotide position 168 was generated from Y chromosome samples using the following forward 5′AGGCACTGGTCAGAATGAAG and reverse primer 5′ AATGGAAAATCAGCAGCTCCCC, respectively. The forward primer was labeled with 6-FAM (emission maximum, 520 nm). A 481-bp fragment encompassing exon 39 of the ATM gene was amplified with a 6-FAM labeled forward, 5′ GCAGTATGTTGAGTTTATGGCA, and unlabeled reverse primer, 5′ GCAACTGTTGGCAACTTTTAT, respectively. It contained either one of the following single-base substitutions: G>A at nucleotide position 60 (IVS 38–83), G>C at 128 (IVS38–15), and G>A at 204 (nucleotide 5557), respectively. Based on the publicly available melting algorithm (Jones et al. 1999), the fragment was predicted to contain five melting domains of 52°C (1–116), 53°C (117–128), 54°C (129–199, 361–431), 55°C (200–220, 315–360), and 56°C (221–314), respectively. Finally, three fragments were generated from the MDR1 gene encompassing the coding and flanking noncoding regions of exon 13 (380 bp; forward, 5′-HEX-ATCTTTCTGATGTTGCCCTTTC; reverse, 5′-NED-CCTTCTTAGGATTTCCCTTCTT), exon 22 (351 bp; forward, 5′-FAM-ACCACTATTTACTCTTGTGCCT; reverse, 5′-GTTCTACCTTAGAGATGTCCCT), and exon 26 (327 bp; forward, 5′-NED-TGCTGAGAACATTGCCTATGGAG; reverse, 5′-ROX-AACACTTTCATCCCTTCCTCACA). The emission maxima of HEX, NED, and ROX are 555 nm, 580 nm, and 605 nm, respectively. Only one labeled primer was used at a time and paired with an unlabeled forward or reverse primer. Exon 13 contained two polymorphisms at nucleotide positions 52 (T>G) and 291 (T>C), respectively, whereas exon 22 harbored one variant site at position 320 (G>A), and exon 26 three variant sites at positions 212 (C>T), 325 (T>G), and 346 (T>C), respectively. Both unlabeled and HPLC purified fluorescent dye-labeled oligonucleotides were obtained from either Applied Biosystems or Life Technologies.
PCR
Amplifications were performed in 50 μL reactions that contained 1 unit of AmpliTaq Gold (Applied Biosystems), 10 mM Tris·HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.1 mM each of the four deoxyribonucleotide triphosphates, 0.2 μM each of forward/reverse primers, and 50 ng of genomic DNA. The thermocycling protocol comprised an initial denaturation at 95°C for 10 min to activate AmpliTaq Gold, 14 cycles of denaturation at 94°C for 20 sec, primer annealing at 63°C–56°C with 0.5°C decrements, and extension at 72°C for 1 min, followed by 20 cycles at 94°C for 20 sec, 56°C for 1 min, and 72°C for 1 min, and a final 5-min extension at 72°C. PCR yields of the Y chromosome alleles were determined semiquantitatively on ethidium bromide-stained agarose gels. For DHPLC analysis, unpurified PCR products of each Y-chromosome allele were mixed at a roughly equimolar ratio. All amplicons were subjected to a 3 min 95°C denaturing step followed by gradual reannealing from 95°C to 65°C over 30 min prior to DHPLC analysis to ensure maximum heteroduplex formation. Only then were the amplicons mixed for multiplex analysis.
DHPLC
The synthesis of the monolithic columns has been described previously (Premstaller et al. 2000). The chromatograph consisted of a degasser (DG-1210, Uniflows Co.), a solvent organizer (WellChrom K-1500, Knauer Co.), a pump (WellChrom K-1001), a laser-induced fluorescence scanner (see below), and a PC-based data system. A tee-piece was used to split the primary flow to control the secondary low-flow stream to the columns.
Two different column ovens were used. The first one was a forced air circulation oven with Peltier cooling that is commercially available from Varian Analytical Instruments under the brand name MISTRAL. The second oven was custom-made and consisted of a brass tube (15 × 1.2 cm i.d., 0.2 mm thick) wrapped with ∼500 × 0.24 mm o.d. nickel-chromium heating wire over a layer of Mylar tape that provided electrical insulation. This construct was wrapped with additional layers of Mylar tape and the ends of the tube were plugged with foam inserts through which the connecting upstream and downstream tubing passed. Temperature control was implemented by using an Omega CN3390 10-channel temperature control module (only four channels were used in this study) and a reading type T thermocouple that was attached to one of the stainless steel unions used to connect the column to the inlet and outlet lines. It is critical that the thermocouple does not touch the brass tube. The temperature control unit was operated in on-off mode with a dead-band of 0.07°C.
A four-position microscale injector (Model 7410, Rheodyne Inc.) with a 1 μL internal sample loop was used for all analyses requiring injection of a single sample. For the loading of four different samples onto four different columns, a custom-made electrically actuated injector with four internal 1 μL sample loops was obtained from Valco Instruments Co.. The injection valve was connected upstream to a stainless steel 1/16 inch manifold (model no. Z4M1, Valco) with one inlet and four outlets.
The composition of the eluents was (A) 100 mM TEAA (Applied Biosystems), 0.1 mM Na4EDTA at pH 7.0, (B) 100 mM TEAA, 0.1 mM Na4EDTA at pH 7.0, 25% acetonitrile (J.T. Baker). The gradient profiles used for the separations are given in the figure legends.
Laser-Induced Fluorescence Detector
The confocal fluorescence capillary array detector was constructed according to the original scanner by Kheterpal et al. (1996) designed for the purpose of capillary array electrophoresis-based DNA sequencing. This scanner collects four-color fluorescence data from up to 25 capillaries in parallel. Briefly, capillaries are mounted on a translation stage driven by a computer controlled Compumotor microstep indexer (Z6104, Parker Hannifin Co.). Excitation beam (488 nm) from an argon ion laser is focused into the capillary array through a microscope objective. Fluorescence from the capillaries is collected by the same objective and fractioned successively passing through four dichroic beam splitters (Omega Optical) with transmission wavelengths (>50% T) of 505 nm, 540 nm, 575 nm, and 595 nm, respectively. Bandpass filters (Omega, 525DF30, 560DF30, 585DF20) are used on the light reflected by each of the first three beam splitters, and a line filter (10LF10–488) after the last splitter to remove plasma lines from the laser. Filtered light in each fraction is then detected by a photomultiplier tube (Photonics Research). Signals are amplified and collected by means of a computer (CIO-DAS1402/16, National Instruments). A computer program synchronizes the movement of the translation stage and data acquisition, allowing each capillary to be scanned at the rate of twice per second.
Acknowledgments
This study was supported by NIH (HG01932) and the Austrian Science Fund (P-14133-PHY). A.P. is the recipient of an Erwin Schrödinger stipend (J1922) awarded by the Austrian Science Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL oefner@genome.stanford.edu; FAX 650-812-1975.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.200401.
REFERENCES
- Bayliss MK, Little D, Mallett DN, Plumb RS. Parallel ultra-high flow-rate liquid chromatography with mass spectrometric detection using a multiplex electrospray source for direct, sensitive determination of pharmaceuticals in plasma at extremely high throughput. Rapid Commun Mass Spectrom. 2000;13:1165–1168. doi: 10.1002/1097-0231(20001115)14:21<2039::AID-RCM130>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bentley A, MacLennan B, Calvo J, Dearolf CR. Targeted recovery of mutations in Drosophila. Genetics. 2000;156:1169–1173. doi: 10.1093/genetics/156.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Feng B, Patel AH, Keller PM, Slemmon JR. Fast characterization of intact proteins using a high-throughput eight-channel parallel liquid chromatography/mass spectrometry system. Rapid Commun Mass Spectrom. 2001;15:821–826. doi: 10.1002/rcm.276. [DOI] [PubMed] [Google Scholar]
- Gao Q, Yeung ES. High-throughput detection of unknown mutations by using multiplexed capillary electrophoresis with poly(vinylpyrrolidone) solution. Anal Chem. 2000;72:2499–2506. doi: 10.1021/ac991362w. [DOI] [PubMed] [Google Scholar]
- Gong X, Yeung ES. An absorption detection approach for multiplexed capillary electrophoresis using a linear photodiode array. Anal Chem. 1999;71:4989–4996. doi: 10.1021/ac9900634. [DOI] [PubMed] [Google Scholar]
- Gross E, Arnold N, Pfeifer K, Bandick K, Kiechle M. Identification of specific BRCA1 and BRCA2 variants by DHPLC. Hum Mutat. 2000;16:345–353. doi: 10.1002/1098-1004(200010)16:4<345::AID-HUMU7>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Hoskins RA, Phan AC, Naeemuddin M, Mapa FA, Ruddy DA, Ryan JJ, Young LM, Wells T, Kopczynski C, Ellis MC. SNP markers for genetic mapping in Drosophila melanogaster. Genome Res. 2001;11:1100–1113. doi: 10.1101/gr.178001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber CG. Micropellicular stationary phases for high-performance liquid chromatography of double-stranded DNA. J Chromatogr. 1998;806:3–30. doi: 10.1016/s0021-9673(97)01124-2. [DOI] [PubMed] [Google Scholar]
- Huber CG, Premstaller A, Xiao W, Oberacher H, Bonn GK, Oefner PJ. Mutation detection by capillary denaturing high-performance liquid chromatography using monolithic columns. J Biochem Biophys Meth. 2001;47:5–19. doi: 10.1016/s0165-022x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- Jones AC, Austin J, Hansen N, Hoogendoorn B, Oefner PJ, Cheadle JP, O'Donovan MC. Optimal temperature selection for mutation detection by denaturing HPLC and comparison to single-stranded conformation polymorphism and heteroduplex analysis. Clin Chem. 1999;45:1133–1140. [PubMed] [Google Scholar]
- Kheterpal I, Scherer JR, Clark SM, Radhakrishnan A, Ju J, Ginther CL, Sensabaugh GF, Mathies RA. DNA sequencing using a four-color confocal fluorescence capillary array scanner. Electrophoresis. 1996;17:1852–1859. doi: 10.1002/elps.1150171209. [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S. Targeted screening for induced mutations. Nat Biotechnol. 2000;18:455–457. doi: 10.1038/74542. [DOI] [PubMed] [Google Scholar]
- Nickerson ML, Warren MB, Zbar B, Schmidt LS. Random mutagenesis-PCR to introduce alterations into defined DNA sequences for validation of SNP and mutation detection methods. Hum Mutat. 2001;17:210–219. doi: 10.1002/humu.6. [DOI] [PubMed] [Google Scholar]
- Oefner PJ, Underhill PA. DNA mutation detection using denaturing high-performance liquid chromatography. In: Dracopoli NC, et al., editors. Current protocols in human genetics. New York: Wiley and Sons; 1998. pp. 7.10.1–7.10.12. [DOI] [PubMed] [Google Scholar]
- Ophoff RA, Terwindt GM, Vergouwe MN, van Eijk R, Oefner PJ, Hoffman SM, Lamerdin JE, Mohrenweiser HW, Bulman DE, Ferrari M, et al. Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell. 1996;87:543–552. doi: 10.1016/s0092-8674(00)81373-2. [DOI] [PubMed] [Google Scholar]
- Premstaller A, Oberacher H, Huber CG. High-performance liquid chromatography-electrospray ionization mass spectrometry of single- and double-stranded nucleic acids using monolithic capillary columns. Anal Chem. 2000;72:4386–4393. doi: 10.1021/ac000283d. [DOI] [PubMed] [Google Scholar]
- Schriml LM, Peterson RJ, Gerrard B, Dean M. Use of denaturing HPLC to map human and murine genes and to validate single-nucleotide polymorphisms. BioTechniques. 2000;28:740–745. doi: 10.2144/00284rr03. [DOI] [PubMed] [Google Scholar]
- Spiegelman JI, Mindrinos MN, Fankhauser C, Richards D, Lutes J, Chory J, Oefner PJ. Cloning of the Arabidopsis RSF1 gene by using a mapping strategy based on high-density DNA arrays and denaturing high-performance liquid chromatography. Plant Cell. 2000a;12:2485–2498. doi: 10.1105/tpc.12.12.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman JI, Mindrinos MN, Oefner PJ. High accuracy DNA sequence variation screening by DHPLC. BioTechniques. 2000b;29:2–7. doi: 10.2144/00295rr04. [DOI] [PubMed] [Google Scholar]
- Thorstenson YR, Shen P, Tusher VG, Davis RW, Chu G, Oefner PJ. Global analysis of ATM polymorphism reveals significant functional constraint. Sequence diversity of the human ATM gene. Am J Hum Genet. 2001;69:396–412. doi: 10.1086/321296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ. Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res. 1997;7:996–1005. doi: 10.1101/gr.7.10.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TMU, Stoppa-Lyonnet D, Fleischmann E, Muhr D, Pagès S, Sandberg T, Caux V, Moeslinger R, Langbauer G, Borg A, Oefner P. Denaturing high performance liquid chromatography (DHPLC) detects reliably BRCA1 and BRCA2 mutations. Genomics. 1999;62:369–376. doi: 10.1006/geno.1999.6026. [DOI] [PubMed] [Google Scholar]
- Wolford JK, Blunt D, Ballecer C, Prochazka M. High-throughput SNP detection by using DNA pooling and denaturing high performance liquid chromatography (DHPLC) Hum Genet. 2000;107:483–487. doi: 10.1007/s004390000396. [DOI] [PubMed] [Google Scholar]
- Xiao W, Oefner PJ. Denaturing high-performance liquid chromatography: A review. Hum Mutat. 2001;17:439–474. doi: 10.1002/humu.1130. [DOI] [PubMed] [Google Scholar]
- Xiao W, Stern D, Jain M, Huber CG, Oefner PJ. Multiplex DNA variation analysis by capillary denaturing high-performance liquid chromatography and laser-induced fluorescence detection. BioTechniques. 2001;30:1332–1338. doi: 10.2144/01306rr01. [DOI] [PubMed] [Google Scholar]