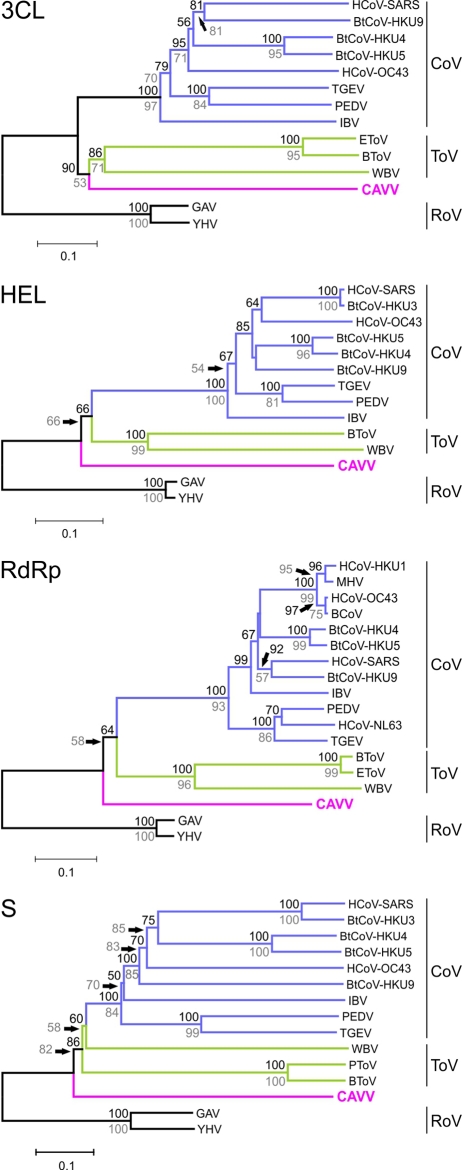
FIG 5 .
Phylogenetic relationship of CAVV to prototype nidovirus strains for selected genomic regions. Phylogenetic analyses were performed using the NJ algorithm, a BLOSUM62 substitution matrix, and no distance correction. Indels were fully deleted. Significance was tested by bootstrap analysis using 1,000 resampling steps as implemented in MEGA 5.0 (66). Bootstrap values are shown above nodes. Analyses were also performed using the maximum-likelihood algorithm in FastML with the same settings. Results of this analysis are not shown because of congruent topologies. Bootstrap support values (1,000 replicates) from maximum-likelihood analysis are shown in grey below nodes.