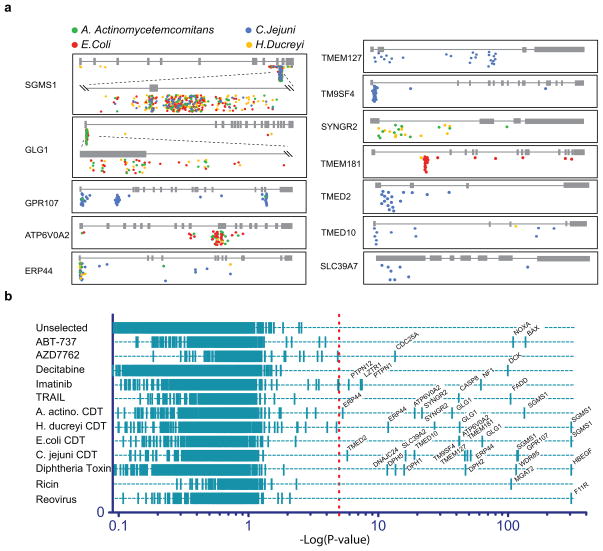
Figure 3. Genes linked to different phenotypes.
A. Gene trap insertions identified in loci essential for CDT intoxication. A color code distinguishes gene trap insertions that were enriched using distinct CDTs: A. actinomycetemcomitans in green, E. coli in red, H. ducreyi in yellow and C. jejuni in blue. B. Loci linked to 12 separate phenotypes. Cells were exposed to small molecule inhibitors of the Bcl2-family (ABT-737), chk1-kinase activity (AZD7762), Bcr-Abl-activity (imatinib), or DNA methylation (decitabine). Additional phenotypic selections were done using biological agents including TRAIL, CDTs, diphtheria toxin, ricin toxin and reovirus. Gene trap insertions in exonic sequences or in the sense orientation of genes were counted per individual selection. Enrichment p-value was calculated for each gene locus by comparing this number to the number of insertions identified in the same locus within the unselected cell population. Each screen resulted in a distinct set of between 1 and 8 genes with high significance. These include already known entry factors used by pathogens such as the entry receptor for diphtheria toxin (HBEGF), the reovirus receptor (F11R) and an enzyme involved in carbohydrate synthesis required for ricin entry (MGAT2). It also includes downstream effectors of kinases for example CDC25A in a screen with a Chk1 inhibitor or PTPN1 and PTPN12 identified by BCR-ABL inhibition using imatinib. Strong resistance against decitabine was observed in cells containing mutations in deoxycytidine kinase (DCK), the rate-limiting kinase for activation of several nucleoside analogs20.