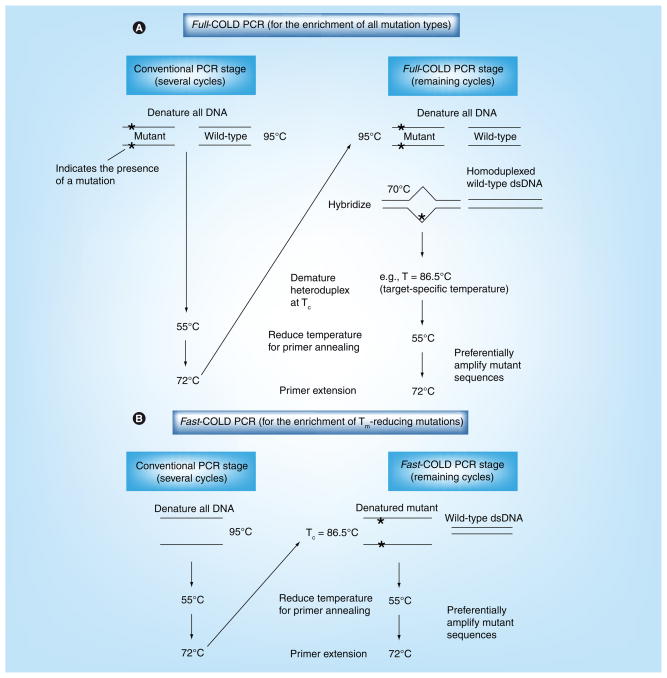
Figure 1. Coamplification at lower denaturation temperature-PCR protocol.
Two original forms of COLD-PCR were developed as full-COLD-PCR (A) and fast-COLD-PCR (B). (A) Full-COLD-PCR has the potential to enrich all possible mutations. Several preliminary rounds of conventional PCR enable an initial increase of the target amplicon(s). After denaturation at approximately 95.0°C (or as defined by the polymerase system), the PCR amplicon(s) are incubated (e.g., 70.0°C for 2–8 min) for re-annealing and hybridization. Hybridization of mutant and wild-type alleles forms heteroduplexed molecules (mismatch-containing) that possess a lower Tm than homoduplexed molecules. The PCR temperature is subsequently increased to the Tc (e.g., Tc = 86.5°C) to preferentially denature the heteroduplexed amplicons. The temperature is reduced for primer annealing (e.g., 55.0°C), and then increased to 72.0°C for primer extension, thus preferentially amplifying the mutation-containing alleles. (B) Fast-COLD-PCR can be performed to enrich mutations with melting temperatures lower than the wild-type amplicon. Denaturation at the Tc (rather than the standard 95.0°C) preferentially denatures the strands containing the lower Tm allele; this generates single-stranded DNA for primer annealing and extension.
COLD: Coamplification at lower denaturation temperature; dsDNA: Double-stranded DNA; Tc: Critical denaturation temperature; Tm: Melting temperature.