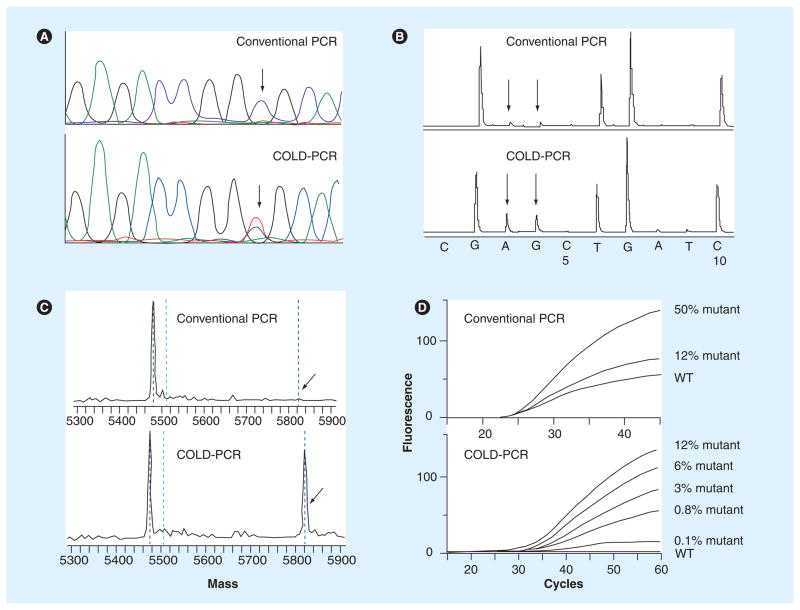
Figure 5. COLD-PCR improves mutation detection in downstream assays.
(A) Sanger sequencing detects low-abundance mutations after COLD-PCR. A 10% abundance of a C>T mutation in WT DNA was amplified by both conventional PCR and COLD-PCR. Sanger sequence chromatograms were evaluated for both approaches, respectively (conventional PCR is presented in the upper panel; COLD-PCR is presented in the lower panel); an approximate sixfold mutation enrichment by COLD-PCR is evident. (B) COLD-PCR improves detection via pyrosequencing. A 33-fold dilution of a G>A mutation in WT DNA was amplified by both conventional and COLD-PCR. The mutant is only visible in the COLD-PCR amplicons. (C) COLD-PCR improves the sensitivity of MALDI-TOF genotyping technologies. A G>A mutation was amplified by both conventional and COLD-PCR, and amplicons were genotyped using MALDI-TOF. The G>A mutation was undetectable in conventional PCR amplicons (upper panel); however, it was detectable in COLD-PCR amplicons (lower panel). (D) COLD-PCR improves the sensitivity of TaqMan® genotyping technologies [35]. Serial dilutions of the human H1975 cell line (containing the T790M mutation in EGF receptor exon 20) in WT DNA were screened with conventional and COLD-PCR TaqMan genotyping for the T790M mutation. Conventional PCR TaqMan genotyping for the T790M mutation is presented in the upper panel; COLD-PCR TaqMan genotyping for the T790M mutation is presented in the lower panel.
COLD: Coamplification at lower denaturation temperature; WT: Wild-type.