Abstract
H11/HspB8 is a functionally distinct small heat shock protein. It causes growth arrest in melanocytes, associated with inhibition of cyclin E/cdk2 and -catenin phosphorylation at the transcriptional activity site Ser552 and is silenced through DNA methylation in 27/35 (77%) melanoma tissues/early cultures. 5'-Aza-2-deoxycytidine (Aza-C) induces melanoma cell death correlated with the levels of H11/HspB8 DNA methylation (p<0.001). In lines with low/moderate H11/HspB8 methylation, PI3-K inhibition increases Aza-C-induced cell death. Aza-C Inhibits growth of melanoma xenografts related to the levels of H11/HspB8 methylation, and a non-methylated/non-TAK1 binding H11/HspB8 mutant confers Aza-C resistance. H11/HspB8 is a potential molecular marker for demethylation therapies.
INTRODUCTION
The incidence of melanoma is increasing worldwide at a faster rate than that of any other cancer[1] and many patients with stage IV disease often die within 2 years with few long term survivors[2]. Therapeutic options are minimal and their efficacy is low, underscoring the need for improved strategies that are based on a better understanding of the molecular mechanisms of tumor development/progression. A complex molecular machinery that provides checks and balances in normal melanocytes, is altered in melanoma through chromosomal deletions, amplifications and gene mutations[3,4]. However, most of these alterations occur at a relatively low frequency and their potential as therapeutic targets is unclear.
Recent interest has focused on the role of epigenetic modifications in melanoma pathogenesis and as a potential therapeutic target. DNA methylation of cytosine residues that are part of a CpG dinucleotide (5'CG3') is the main epigenetic modification in cancer[5]. The human genome contains regions of unmethylated segments interspersed by methylated regions known as CpG islands. Approximately half of all the human genes have CpG islands, and these are present on both housekeeping genes and genes with tissue-specific expression patterns. DNA methylation is an important regulator of gene transcription, and its role in carcinogenesis is of considerable interest. Transcriptional inactivation of tumor suppressor genes by DNA hypermethylation is documented in many human cancers[6] and it has been suggested to play a central role in melanoma progression[7]. Global genome analyses identified a large number of hypermethylated genes in melanoma. However, the methylation patterns differed in various melanoma lines, and the ability of demethylation to cause melanoma cell death as well as the involved molecular target(s), remain unknown[8–13]. As oncology moves forward into the field of personalized therapies these limitations are particularly troublesome because molecular and prognostic markers are crucial aspects of patient selection for individual targeted treatments.
The H11 gene (Entrez ID # 26353) is also known as Hsp22 and HspB8. It was originally cloned in our laboratory and shown to map at a chromosome site (12q24) with frequently reported allelic deletions in malignant neoplasms[14–16]. Although the H11/HspB8 protein retains the α-crystallin motif characteristic of small heat shock proteins (Hsp), it differs from canonical family members in many biological and functional properties, including association with the plasma membrane and presence of nuclear export sequences, failure to translocate to the nucleus upon heat shock, and the presence of intrinsic protein kinase activity[14,15,16–22]. The H11/HspB8 promoter is rich in CpG islands (Supplementary Information, Fig S1) and we have previously shown that H11/HspB8 is silenced in some melanoma cell lines in which its forced expression triggers apoptosis[20,21,23]. The studies described in this report were designed to determine the function of H11/HspB8 in normal melanocytes, examine its methylation status in melanoma tissues and xenografts and evaluate the potential of H11/HspB8 as a marker for demethylation-based therapies.
MATERIAL AND METHODS
Melanoma tissues and early and late passage cultures
Because previous studies had shown that the methylation status of various genes can change with cell passage[24], we studied three types of cell preparations: (i) tissue cell suspensions, (ii) early passage in vitro cultures and (iii) late passage in vitro cultures. The tissue cell suspensions were the gift Dr. George Elias (Franklin Square Hospital, Baltimore, MD). They had been prepared from 27 metastatic melanoma tissues (stage IV) collected at surgery and given 4 serial passages in Iscove's modified Dulbecco medium with 1 μg/ml gentamycin and 10% fetal bovine serum (FBS) (Gemini Bioproducts, Calabasos, CA) . Thy ha been cryopreserved in liquid nitrogen at −80°C before we received them. Flow cytometry done at the time that the cell suspensions were received showed that 85±8% of the cells stained with antibodies to melanoma markers MART1/MelanA, S100 and HMB45[25], indicating that they had a relatively high degree of homogeneity for melanoma cells. These suspensions were used in methylation assays without further in vitro passage. The identity of the small percentage of cells negative for the melanoma markers is unclear and likely represents contaminating cells, possibly fibroblasts. The second type of cell preparation used in these studies consisted of 8 early passage in vitro cultures established from metastatic melanoma tissues. They were also the gift of Dr. George Elias, and they contained 91–97% MART1+ cells as determined by flow cytometry. The cultures were grown in Iscove's modified Dulbecco medium with 1 μg/ml gentamycin and 10% FBS for 12–17 passages before study. The late passage in vitro cultures that we studied included established melanoma cell lines A2058, A375, MeWo, SKMEL-2, HT144 and SKMEL-28 obtained from the American Type Culture Collection (Manassas, VA) and LM cells, which were the gift of Dr. Joseph Sinkovics (University of South Florida, Tampa, FL). SKMEL-28 cells were grown in Eagle's minimal essential medium (EMEM) with 1mM sodium pyruvate, 0.1mM nonessential amino acids and 10% FBS. All the other cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) with 4.5g/L glucose, 1500mg/ml sodium bicarbonate, 4mM glutamine and 10% FBS. LM cells were established from a metastatic melanoma and grown in RPMI 1640 with 10% FBS. HMB45 staining was seen in 97% of the cells. LM cultures were studied at passages 50–72. A2058 and SKMEL-2 cells stably transfected with tet-inducible H11/HspB8 were previously described[23]. Normal skin tissue (NS#3) and adult normal human melanocytes (NHM) were purchased from Cascade Biologicals (Portland, Ore). NHM were studied at passages 2–5. Growth medium, provided by the company consisted of medium 154 supplemented with 3ng/ml basic fibroblast growth factor, 0.3% bovine pituitary extract, 3 μg/ml heparin, 0.18 μg/ml hydrocortisone, 5 μg/ml insulin, 5 μg/ml transferrin, 10nM endothelin-1 (human melanocyte supplement 2). Human embryonic kidney (HEK-293) cells were obtained from the American Type Culture Collection and grown in EMEM with 1 mM nonessential amino acids, 1% sodium pyruvate, and 10% FBS
Antibodies and chemical reagents
The generation and specificity of the rabbit polyclonal antibody to H11/HspB8 (recognizes amino acids 181–194) were previously described[14]. Antibodies to Cdk2, Cyclin E, TAK1, β-catenin, caspase-3 (recognizes both the zymogen and its cleavage products), GST and actin, were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies for β-catenin phosphorylated on Ser552 (pβ-catenin-Ser552), Ser675 (pβ-catenin-Ser675) or Thr41/Ser45 (pβ-catenin-Thr41/Ser45) were from Cell Signaling Technologies (Danvers, MA). 5-Aza 2'-deoxycytidine (Aza-C) was purchased from Sigma Chemicals (St. Louis, MO, USA), doxycycline (Dox) from Clontech (Mountainview CA) and LY294002 from Cell Signaling Technology (Danvers, MA). The WST-1 Quick Cell Proliferation kit was purchased from Biovision Research Products (Mountainview CA) and used as per manufacturer's instructions.
Methylation-Specific PCR (MSP) and quantitative real time MSP (MSQP)
Genomic DNA was extracted with the Wizard Genomic DNA purification kit (Promega, Madison, Wisc., USA) and sodium-bisulfite modified with the EZ DNA Methylation kit (Zymo Research Corp, Orange, CA., USA), both according to manufacturers' instructions. The primers for the MSP assay were 5-GGGAAAGGTAGGATACGATA CGTTT-3 (sense) and 5-ACTAAAACCGCCCTCCCCGAACGA-3 (antisense). For the PCR reaction, bisulfite converted DNA was amplified using a Gene Amp Gold PCR kit (Applied Biosystems, Carlsbad, CA) with 20 pmol of each primer in a 50μl reaction volume for 35 cycles (95°C for 1 min, 58°C for 1 min, and 72°C for 1 min). Reaction products were separated by electrophoresis on 1.7% agarose gels and detected by staining with 0.5 μg/ml ethidium bromide. The MSQP assay was done as described by Martinez-Galan et al.,[26] using the QuantiTest SYBR green PCR Kit (Qiagen, Valencia CA). Each PCR reaction (20μl) contained 200 ng of the bisulfite-converted DNA, 2×QuantiTect SYBR Green PCR Master Mix (10μl) and 0.2μM each sense (5'-GGGAAAGGTAGGATACGATACGTTT-3') and antisense (5'-TCCAAATTCAACCGA CTCTCCGA-3') primers (position −224bp to −21bp relative to the transcription start site) which will amplify a DNA product that contains 9 CpG sites (Supplementary Information, Fig S1). Real-time PCR conditions were 95°C for 15 min followed by 41 cycles of 94°C for 15s, 55 °C for 25s and 72 °C for 25s and one additional cycle of 76 °C for 10s with data acquisition after each cycle. Duplicate PCR reactions were run using the LightCycler 2.0 Instrument (Roche Applied Science, Germany) and data analysis was with the LightCycler software 3.0. Cycle threshold (Ct) values, defined as the cycle number at which the fluorescence-generated signal crosses an arbitrary but defined threshold within the exponential phase of the reaction, were converted to methylation units from a standard curve generated with serial dilutions of the CpGenome™ Universal methylated DNA (Millipore, Temucala CA). Relative methylation units are expressed as the dilution factor[26].
Quantitative Real time RT-PCR (QRT-PCR)
Total RNA was isolated with the RNeasy Mini Kit (Qiagen) and cDNA was generated by reverse transcription using the High-Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA), both according to the manufacturers' instructions. The Real time QuantiTect SYBR green PCR Kit (Qiagen) was used for quantitative amplification of the cDNA. Each assay mixture contained 1μl cDNA and 0.3μM sense and antisense primers. The H11/HspB8 primers were 5'-CCATGGCTGACGGTCAGATGCCCTTCTCCT-3' (sense) and (5'-TCCATGCCAAAGCCATCATCCAGCAG-3 (antisense). The primers for human β-actin, (used for data normalization), were 5'-CATGTACGTTGCTATCCAGGC-3' (sense) and 5-CTCCTTAATGTCACGCACGAT-3' (antisense). Real-time PCR conditions were 95°C for 15 min followed by 40 cycles of 94°C for 15s, 64°C for 30s and 72°C for 30s. All the reactions were done in duplicate using the LightCycler 2.0 Instrument (Roche) and data analysis was with the LightCycler software 3.0 to generate average Ct values. Results are expressed as ΔCt = Ct for H11/HspB8 – Ct for β-actin. Low ΔCt values indicate high expression[27].
H11/HspB8 DNA sequencing
cDNA was PCR amplified with H11/HspB8 primers 5'-ACCATGGCTGACGGTCAGATGCCCTTC-3' (sense) and 5'-ATCTCAGGT ACAGGTGACTTCCTGGC-3' (antisense) (0.8 μM) using the REDTaq™ kit (Sigma) as per manufacturer's instructions. The PCR program was 5 minutes at 94° C for 1 cycle, followed by 94°C for 30 seconds, 62°C for 30 seconds, and 72°C for 30 seconds for 35 cycles, with a final extension at 72°C for 6 minutes. The products were purified with the QIAquick PCR Purification Kit (Qiagen) and sequenced by the University of Maryland Biopolymer Core Facility.
H11/HspB8 antisense oligonucleotide
The sequence and specificity of the phosphorothioate oligodeoxynucleotides (ODN) complementary to the H11/HspB8 translation initiation site [antisense ODN (aODN); 5'-CCGTCAGCCATGGTGG-3'] and the sense ODN (sODN; 5'-CCACCATGGCTGACGG-3') were previously described[14].
Immunoprecipitation and immunoblotting
Cells were lysed with immunoprecipitation buffer with protease and phosphatase inhibitor cocktails (Sigma-Aldrich) and protein extracts were assayed by immunoblotting or immunoprecipitation/immunoblotting, as previously described[14,15,20,23]. Densitometry used the Bio-Rad GS-700 imaging densitometer. Pixel density was determined with Multi Analyst software (Bio-Rad, Hercules, CA) and the results are expressed as mean densitometric units ± SD for three independent experiments normalized to actin.
Construction of GST-H11/HspB8 fusion protein and protein kinase (PK) assays
The GST-H11/HspB8 fusion protein was generated as previously described[14] using the prokaryotic expression vector pGEX-4T-3 (Amersham Pharmacia Biotech). Transformation was in the protease-deficient E. coli BL21 and expression was induced by treatment (4h; 37°C) with 0.1mM isopropyl-1-thio-β-D-galactopyranoside (IPTG). Glutathione-Sepharose 4B beads coated with bacterial extracts induced to express GSTH11/Hsp8 or GST alone, were used in PK assays with purified GST-β-catenin or GST proteins (2μg) (SignalChem Richmond, British Columbia) as substrates. Each assay mixture contained 20mM Tris-HCl pH 7.4, 5mM MgCl2, 2mM MnCl2, and 20μCi [32P]-γ-ATP (3000Ci/mM; Perkin Elmer, Waltham, MA) and incubation was at 30°C for 15 min. Proteins were separated by SDS-PAGE, transferred to PVDF membranes and phosphorylation was verified by exposure to X-ray film followed by immunoblotting with antibodies to β-catenin or GST.
In vivo studies
The Animal Care and Use committee of the University of Maryland School of Medicine approved all the described studies. Six-eight week old female nude mice (Balb/c nu/nu) were obtained from Charles River Laboratories (Wilmington, MA). To establish subcutaneous melanoma xenograft models, nude mice were given melanoma cells (5 × 106 in 100μl) by subcutaneous injection into both hind flanks. Animals were randomly divided into two groups and left untreated or given ip injections of Aza-C (5mg/kg) (3 doses at 3hr intervals) on days 6 and 15 after the tumors became palpable (200 mm3 in volume). Each treatment group consisted of 10 tumors. Minimum and maximum perpendicular tumor axes were measured with microcalipers and tumor volume was calculated using the formula: volume=[(length × width2)/2]. Animals were maintained in pathogen-free conditions and were euthanized when their tumors reached 1.5 cm in any one direction. Tissues were collected at that time.
Statistics
Pearson's correlation coefficients were calculated using SAS software. Analysis of variance (ANOVA) was performed with SigmaStat version 3.1 for Windows (Systat Software, Point Richmond, CA). Tumor volumes were compared between untreated and treated groups by pairwise two-way ANOVA followed by the Tukey's honestly significant difference test.
RESULTS
H11/HspB8 is a cell cycle regulator in normal melanocytes
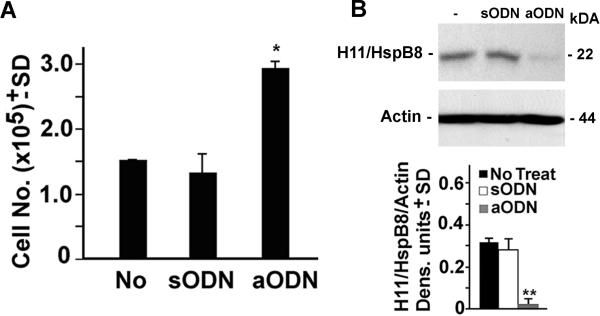
To examine the function of H11/HspB8 in normal melanocytes, NHM cultures were grown (37° C; 3d) in triplicate in the absence or presence of 20μM of H11/HspB8 antisense oligonucleotide (aODN) or sense oligonucleotide (sODN) (used as control) and examined for cell growth by trypan blue staining. The effect of the ODNs on H11/HspB8 expression was confirmed by immunoblotting of protein extracts from duplicate cultures. Relative to the untreated cultures, treatment with aODN caused a significant increase in the cell numbers (p < 0.01) that was associated with inhibition of H11/HspB8 expression. H11/HspB8 expression was not inhibited by sODN and it did not alter the cell number (Fig. 1). Flow cytometry, done as previously described[14,15,23], showed that the aODN-treated cultures had a decreased proportion of cells in the G1 phase (45–47%) and an increased proportion in the S phase (42–44%) relative to the untreated and sODN treated cultures (58–61% and 21–23% cells in G1 and S phases, respectively). The proportion of cells in G2 was slightly lower in the aODN than untreated or sODN-treated cultures (9–13% and 16–21%, respectively), indicating that H11/HspB8 inhibits growth at the G1/S phase of the cell cycle.
Fig. 1. H11/HspB8 causes growth arrest in normal melanocytes.
(A) Viable cell counts in NHM cultures treated or not with H11/HspB8 aODN or sODN (20μM; 3 days). Data are expressed as cell number × 105 ± SD. (B) Protein extracts from similarly treated replicate cultures were immunoblotted with antibody to H11/HspB8. Blots were stripped and re-probed with actin antibody and data are expressed as H11/HspB8 densitometric units normalized to actin ± SD. Representative molecular weights are shown on the right. * p< 0.01; ** p< 0.001 vs No treat, by ANOVA.
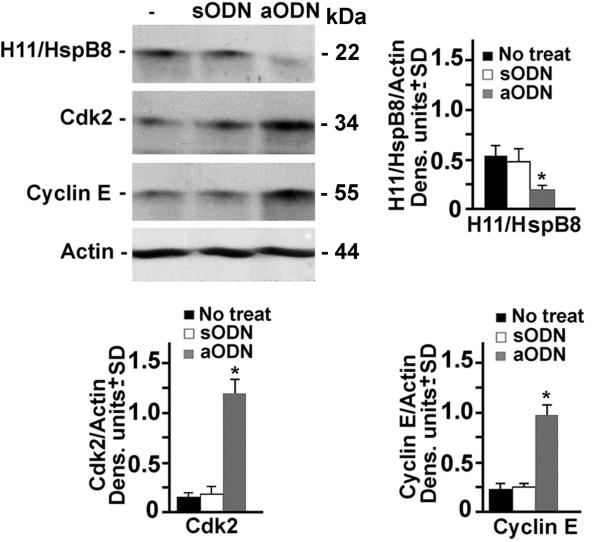
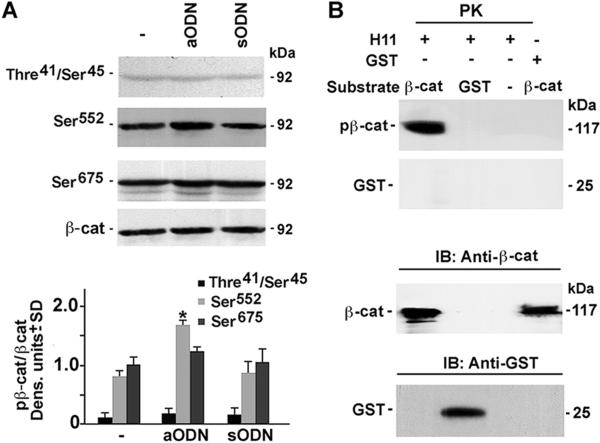
The levels of Cyclin E and Cdk2, that are required for G1/S transition[28,29], were significantly higher in aODN-treated NHM than in their untreated or sODN-treated counterparts, indicating that H11/HspB8 inhibits the Cyclin E/Cdk2 complex (Fig. 2). This inhibition was accompanied by a significant increase in the levels of β-catenin phosphorylated at Ser552 (pβ-catenin-Ser552), a site that is required for nuclear translocation and transcriptional activity[30,31], but not by β-catenin phosphorylated at two other sites (Ser675 and Thr41/Ser45) that have distinct functions[32–34] (Fig. 3A). Because β-catenin is responsible for Cyclin E expression[31,35], the data suggest that H11/HspB8 inhibits the Cyclin E/Cdk2 complex through its ability to interfere with the phosphorylation of β-catenin at Ser552. While the mechanism whereby H11/HspB8 specifically affects Ser552 is still unclear, we infer that it is related to β-catenin phosphorylation at other sites [36], because in vitro kinase assays with GST-H11/HspB8 coated glutathione-sepharose beads indicated that it phosphorylates purified GST-β-catenin protein used as substrate, but phosphorylation was not seen for GST alone (Fig. 3B). Collectively, the data indicate that H11/HspB8 is a melanocyte growth inhibitor.
Fig. 2. H11/HspB8 inhibits Cyclin E/Cdk2 in NHM.
Protein extracts from NHM cultures treated or not with H11/HspB8 aODN or sODN as in Fig. 1, were immunoblotted with antibody to H11/HspB8. Blots were sequentially stripped and re-probed with antibodies to Cdk2, Cyclin E or actin. Data were quantified by densitometric scanning and results are expressed as densitometric units normalized to actin ± SD. Representative molecular weights are shown on the right. * p< 0.001 vs No treat, by ANOVA.
Fig. 3. H11/HspB8 inhibits β-catenin phosphorylation at Ser552 in normal melanocytes.
(A) Extracts of NHM treated or not with H11/HspB8 aODN or sODN as in Fig. 1 were immunoblotted with antibody to pβ-cateninThre41/Ser45. Blots were sequentially stripped and reprobed with antibodies to pβ-catenin Ser552, pβ-catenin Ser675 or total β-catenin. Data were quantified by densitometric scanning and results are expressed as pβ-catenin/-catenin densitometric units ± SD. (B) In vitro kinase assay with purified GST--cateni n (βcat) or GST proteins as substrates and glutathione-Sepharose beads coated with GST-H11/HspB8 or GST. Products were subjected to polyacrylamide gel electrophoresis and transferred to PVDF membranes. Protein identity was confirmed by immunoblotting with antibodies to catenin or GST. Representative molecular weights are shown on the right. Protein kinase (PK); immunoblotting (IB); * p<0.001 vs No treat, by ANOVA.
H11/HspB8 DNA methylation in cell suspensions from melanoma tissues is inversely related to gene expression
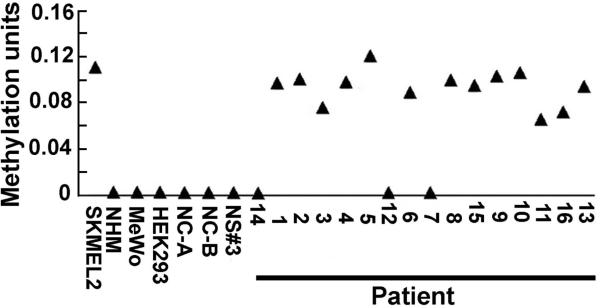
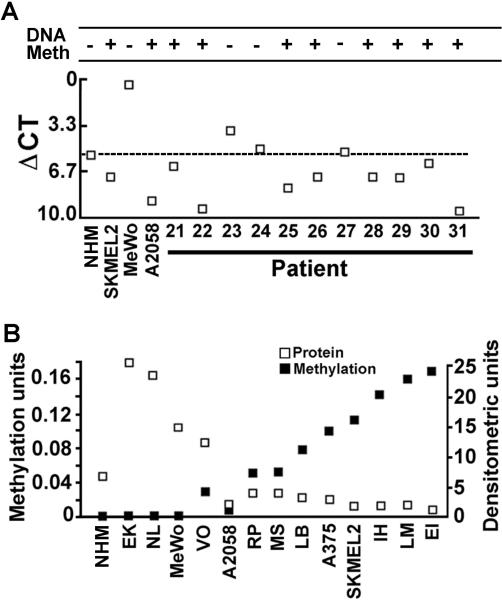
We used cell suspensions from melanoma tissues shown to contain 85±8% cells that expressed melanoma markers MART1/MelanA, HMB45 and S100, as described in Materials and Methods. To examine the incidence and levels of H11/HspB8 methylation in these suspensions, the DNA was assayed by MSQP, as described in Materials and Methods. CpGGenome™ Universal Unmethylated human genomic DNA (NC-A and NC-B) (Millipore) and DNA from NHM cultures and normal skin biopsies were used as negative controls. HEK293 cells, and melanoma cell lines SKMEL-2 and MeWo that are respectively sensitive or resistant to Aza-C-induced cell death[23], served as additional controls. H11/HspB8 DNA was methylated in cell suspensions from 13/16 (81%) melanoma tissues. In all but 3 of these, the levels of methylation were similar to those seen for SKMEL-2 cells and ranged between 0.09 and 0.12 units. Three tissues had lower methylation levels (0.06–0.075 units). Methylation was not seen in MeWo cells, which are Aza-C resistant, NC-A, NC-B, NHM, normal skin tissues and HEK293 cells (Fig. 4), but it was also seen in cell suspensions from 8/11 (73%) melanoma tissues assayed by MSP (Fig. 5A). The data indicate that H11/HspB8 methylation is a relatively common [21/27 (78%)] melanoma-specific event, but the levels of methylation are tissue-specific.
Fig. 4. H11/HspB8 DNA is methylated in melanoma tissues.
Bisulfite treated DNA from single cell suspensions of melanoma tissues prepared as described in Materials and Methods was assayed by MSQP alongside DNA from melanoma cell lines SKMEL-2 and MeWo, HEK293 cells, normal human melanocytes (NHM), normal skin tissues (NS3#) and CpGenome™ Universal Unmethylated DNA (NC-A, NC-B). Data are expressed as methylation units calculated as described in Materials and Methods.
Fig. 5. H11/HspB8 methylation is inversely related to gene expression.
(A) Bisulfite treated DNA and mRNA isolated from cell suspensions of metastatic melanoma tissues, normal human melanocyte cultures (NHM) and melanoma cell lines SKMEL-2, MeWo and A2058 were assayed by MSP and QRT-PCR, respectively. MSP results are expressed as +/− based on the detection of methylated PCR products in 1.7% agarose gels. Data for QRT- PCR are expressed as ΔCt values calculated as described in Materials and Methods. Low ΔCt values indicate high expression. The ΔCt value for NHM (5.4) was used as cut-off point (dotted line) to assess the ΔCt values for the melanoma tissues and they are shown relative to the MSP results. (B) Bisulfite treated DNA from early passage melanoma cultures EK, NL, VO, RP, MS, LB, IH, and EI, established melanoma cell lines MeWo, A2058, A375, SKMEL2, LM and NHM was analyzed by MSQP and gene expression was determined by immunoprecipitation/immunoblotting with H11/ HspB8 antibody. Data are expressed as methylation and densitometric units, respectively. The inverse correlation between H11/HspB8 methylation and gene expression is statistically significant (p< 0.05 by Pearson's correlation coefficient).
Two series of experiments were done to examine the relationship between H11/HspB8 methylation and gene expression. First, the cell suspensions from 11 melanoma tissues examined for H11/HspB8 methylation by MSP, were analyzed for H11/HspB8 mRNA levels by QRT-PCR. Melanoma cell lines SKMEL-2, A2058, and MeWo that have different levels of H11/HspB8 methylation (Fig. 4,5B) and NHM were studied in parallel and served as controls. mRNA levels correlated with DNA methylation, such that all the melanoma cell suspensions with methylated H11/HspB8 DNA had mRNA levels lower than those in NHM (ΔCt = 5.4), but 3 melanoma tissues with unmethylated H11/HspB8 DNA had mRNA levels similar or only slightly higher than the NHM. High mRNA levels were also seen in MeWo cells in which H11/HspB8 is not methylated but not in SKMEL-2 and A2058 cells, in which H11/HspB8 is methylated (Fig. 5A).
Since the MSP assay does not accurately reflect the levels of DNA methylation, a second series of experiments examined DNA methylation by MSQP, which is a very sensitive and quantitative method. Early passage melanoma cultures and cell lines MeWo, A375, A2058, LM, and SKMEL-2 were also examined by MSQP and the levels of methylated DNA were compared to protein expression determined by immunoprecipitation/immunoblotting. NHM cultures were studied in parallel and served as control. H11/HspB8 was methylated in 6/8 early passage cultures (75%) and 4/5 (80%) cell lines. These proportions are similar to those seen for the melanoma tissues, indicating that methylation is not an artifact resulting from cell line establishment. Cultures with moderate/high methylation units (> 0.05) had low expression levels (< 10 densitometric units) and those with low methylation units (<0.05) had high levels of gene expression (>10 densitometric units). Expression was seen in NHM in which H11/HspB8 is not methylated (Fig. 5B). The inverse correlation between the levels of H11/HspB8 methylation and gene expression is statistically significant (p< 0.05 by Pearson's correlation coefficient).
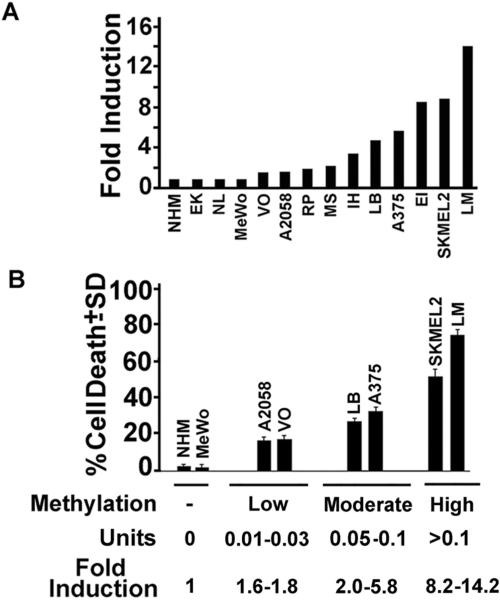
H11/HspB8 DNA methylation correlates with Aza-C induced H11/HspB8 expression and melanoma cell death
Having seen that the levels of H11/HspB8 methylation differ in various melanoma lines, we wanted to know whether they reflect the extent of demethylation-induced cell death. Duplicates of the cultures examined for H11/HspB8 methylation (Fig. 5B) were treated with Aza-C at a relatively low dose (2μM)[10,12], and examined for cell death by the WST-1 assay. Duplicate cultures were assayed for H11/HspB8 expression by immunoprecipitation/immunoblotting and the results are expressed as fold-induction, defined as H11/HspB8 densitometric units in Aza-C treated/untreated cultures. Restored expression was seen in all the cultures with methylated H11/HspB8, but it covered a relatively wide range (1.6 – 14.2-fold) (Fig. 6A). Cell death correlated with the levels of DNA methylation and fold-induction (p<0.001 by Pearson's correlation coefficient). As summarized in Fig. 6B, the lines fell into four distinct groups. Melanoma lines with unmethylated H11/HspB8 and NHM were negative for both H11/HspB8 induction and cell death. Low/moderate induction levels (≤ 5.8 fold) were seen in lines in which DNA methylation was low/moderate (0.01–0.1 units) and this was associated with low/moderate levels of cell killing (16.5±1.7% to 30±2.4%). Lines with strongly methylated H11/HspB8 DNA (> 0.1 units) had high levels of induction (>5.8-fold) and this was associated with robust cell death (51±4.7 to 74.8± 2.4%). The threshold of H11/HspB8 DNA methylation levels for Aza-C induced expression (>1 fold) was 0.01 methylation units.
Fig. 6. Aza-C induced melanoma cell death correlates with H11/HspB8 DNA methylation.
(A) Protein extracts from early passage melanoma cultures, NHM and melanoma cell lines untreated or treated with Aza-C (2μM) were immunoprecipitated immunoblotted with H11/HspB8 antibody. Data are expressed as fold induction (densitometric units Aza-C treated/untreated cells). (B) Melanoma cultures treated with Aza-C as in (A) were assayed for cell death by the WST-1 assay and results are expressed as % cell death ± SD. Representative lines falling into one of the 4 identified methylation groups (none, low, moderate, high) are shown together with the range of methylation units and fold-induction calculated as in (A).
To confirm that Aza-C mediated cell death was due to H11/HspB8 expression, duplicate cultures of SKMEL-2 cells were treated or not with Aza-C (2μM; 4d) in the presence or absence of 20μM H11/HspB8-specific aODN or sODN and assayed for apoptosis by TUNEL. The percentage of Aza-C induced TUNEL+ cells was significantly decreased in cultures given aODN, but not sODN and this was associated with aODN-mediated inhibition of H11/HspB8 expression, as determined by immunoblotting (Supplementary Information Fig. S2). The data support previous findings that forced H11/HspB8 expression causes melanoma cell death[20,23] and indicate that the extent of Aza-C induced cell death correlates with the levels of H11/HspB8 methylation. They suggest that H11/HspB8 methylation is a potential marker for the therapeutic efficacy of Aza-C.
High levels of Aza-C-restored H11/HspB8 expression overcome the cell survival functions of the constitutively activated PI3-K/Akt survival pathway
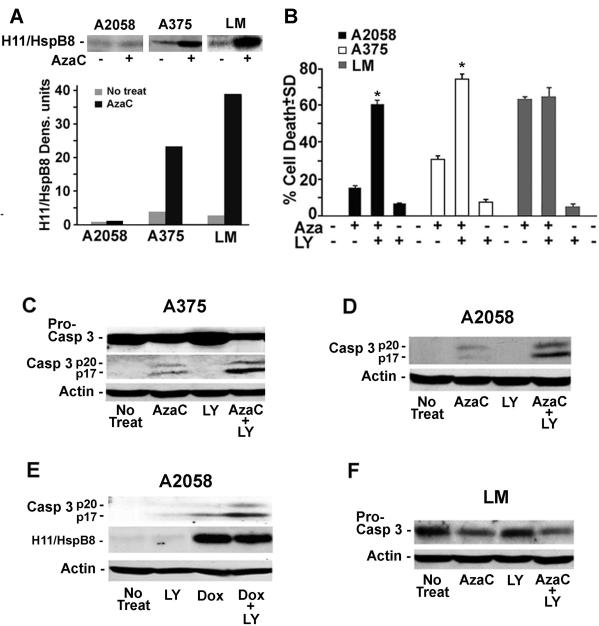
Having seen that Aza-C-induced cell death correlates with the levels of H11/HspB8 DNA methylation/fold-induction, we considered the possibility that the high levels of H11/HspB8 are required in order to overcome the cell survival functions of the PI3-K/Akt pathway that is well known to be constitutively activated in human melanoma[37,38]. Activation of this pathway results in increased levels of phosphorylated (active) Akt[39,40] and is known to cause resistance to chemotherapy[41]. To test the interpretation that high levels of H11/HspB8 are required in order to overcome this cell survival function, we asked whether the levels of Aza-C-induced cell death are differentially affected by PI3-K inhibition in melanoma lines with distinct levels of H11/HspB8 methylation. We used A2058, A375 and LM cells which have significantly increased [3–4-fold] levels of activated (phosphorylated) AKT as compared to NHEM (data not shown) and respectively display low, moderate and high levels of H11/HspB8 DNA methylation (Fig. 5B) and Aza-C-mediated restored expression (1.2 ± 0.1, 23.2 ± 1.7, and 39 ± 2 densitometric units for A2058, A375 and LM, respectively) (Fig. 7A). The cultures were treated with Aza-C (2μM; 4d), the PI3-K inhibitor LY294002 (50μM, 24h) or Aza-C + LY294002 (the latter given for the last 24h of Aza-C treatment) and examined for cell death by the WST-1 assay. H11/HspB8 expression and caspase-3 activation were studied in parallel, by immunoblotting.
Fig. 7. High levels of Aza-C-restored H11/HspB8 expression override the constitutively activated PI3-K/Akt survival pathway.
(A) Extracts of A2058, A375 and LM cells untreated or treated with Aza-C (2μM; 4d) were immunoprecipitated/immunoblotted with antibody to H11/HspB8. Data were quantified by densitometric scanning, and results are expressed as densitometric units. (B) WST-1 assay of A2058, A375 and LM cells untreated or treated with Aza-C (2μM; 4d), LY294002 (50μM; 24h) or Aza-C + LY294002, the latter given for the last 24 h of Aza-C treatment. Data are expressed as % cell death ± SD. * p < 0.001 vs Aza-C by ANOVA. (C,D) Extracts of A375 (C), and A2058 (D) cells treated as in (B) were immunoblotted with caspase-3 antibody (recognizes the procaspase and activated caspase-3). Blots were stripped and re-probed with actin antibody. (E) Extracts of A2058 cells, stably transfected with tet-inducible H11/HspB8, untreated or treated with Dox (5μg/ml; 3d), LY294002 (50μM; 24h), or Dox + LY294002, the latter given for the last 24h of Dox treatment, were immunoblotted with caspase-3 antibody. Blots were stripped and re-probed with antibodies to H11/HspB8 followed by actin. (F) Extracts of LM cells treated as in (B) were immunoblotted with antibodies to caspase-3. ). Blots were stripped and re-probed with actin antibody
The levels of Aza-C induced cell death were directly related to those of H11/HspB8 methylation and fold-induction (15 ± 2.2, 30 ± 2.4 and 62 ± 1.8 % for A2058, A375 and LM cells respectively). Addition of LY294002 caused a dramatic increase in the death of A2058 and A375 (60 ± 2.4 and 75 ± 2.9 %, respectively), but not LM (63 ± 4.5 %) cells, and LY294002 alone had no effect on cell viability (5–15 % cell death) (Fig. 7B). The increased death of A2058 and A375 cells treated with Aza-C+LY294002 is due to enhanced caspase-3 activation, as evidenced by higher levels of activated caspase-3 and lower levels of procaspase-3 than those seen for each drug alone (Fig. 7C,D) and similar results were obtained for A2058 cells stably transfected with tet-inducible H11/HspB and treated with Dox+LY294002 (Fig. 7E). LY294002 did not increase the levels of Aza-C activated caspase in LM cells (Fig.7F). Collectively, the data indicate that Aza-C causes melanoma cell apoptosis the levels of which correlate with those of H11/HspB8 methylation, apparently related to the ability to overcome the cell survival functions of the activated PI3-K/Ak pathway. The H11/HspB8 dose-dependence of the Aza-C-induced melanoma cell death supports the interpretation that H11/HspB8 is one molecular marker for Aza-C treatment.
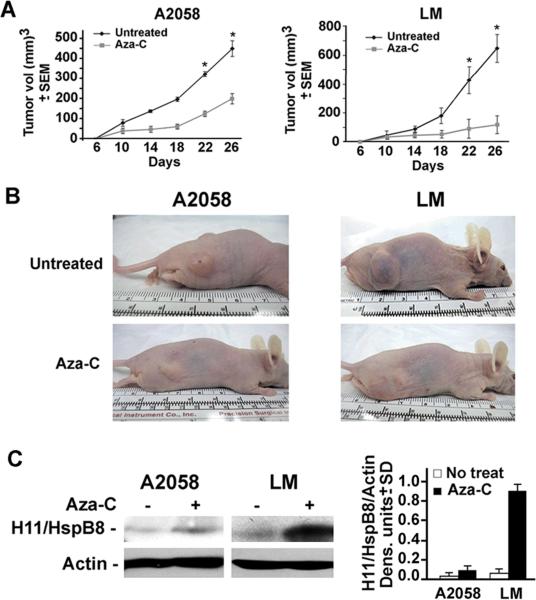
The levels of H11/HspB8 DNA methylation correlate with Aza-C-mediated inhibition of melanoma tumor growth
To examine the relationship between H11/HspB8 methylation and tumor growth inhibition by Aza-C treatment, we used LM and A2058 cells that differ in their sensitivity to Aza-C in culture. Xenografts were established in Balb/c nude (Nu/Nu) mice by subcutaneous injection into both hind flanks. The animals were randomly divided into two groups that were left untreated or given ip injections of Aza-C (5mg/kg) (3 doses at 3hr intervals) on days 6 and 15 after the tumors became palpable and tumor volume was calculated as described in Materials and Methods. Each treatment group consisted of 10 tumors. The untreated xenografts evidenced time-dependent growth reaching maximal levels (651 ± 95.3 and 449.4 ± 43 mm3 for LM and A2058 cells, respectively) at day 26. Aza-C treatment caused a significant decrease in the growth of both tumors (p<0.001 by pairwise two-way ANOVA followed by Tukey post-hoc test). Inhibition was somewhat greater for LM than A2058 xenografts, with respective volumes of 120 ± 61.2 mm3 and 200 ± 25.2 mm3 on day 26 post-treatment. These values respectively represent a 5.4 fold and 2.2 fold inhibition of tumor growth (Fig 8A,B) and correspond to the higher levels of H11/HspB8 methylation (Fig. 5B) and Aza-C- restored expression in LM than A2058 cells (0.88 and 0.086 H11/HspB8/actin densitometric units respectively) (Fig. 8C). The data underscore the critical role of H11/HspB8 in Aza-C induced tumor growth inhibition and its promise as a potential molecular marker for demethylation-based therapeutic efficacy. Studies designed to test this interpretation are ongoing.
Fig. 8. Aza-C-mediated inhibition of melanoma tumor growth correlates with the levels of H11/HspB8 DNA methylation/restored expression.
(A) Nude mice were given A2058 or LM melanoma cells (5 × 106 in 100μl) by subcutaneous injection into both hind flanks and animals randomly divided into two groups were left untreated or given ip injections of Aza-C (5mg/kg) (3 doses at 3hr intervals) on days 6 and 15 after the tumors became palpable (200 mm3 in volume). Each group consisted of 10 tumors. Tumor volume was calculated as described in Materials and Methods. (* p < 0.001). (B) Mice were photographed on day 26,and representative untreated and Aza-C treated animals bearing A2058 or LM tumors are shown. (C) Extracts from representative untreated and Aza-C treated A2058 and LM tumors were immunoblotted with H11/HspB8 antibody. Blots were stripped and re-probed with actin antibody and data are expressed as H11/HspB8 densitometric units normalized to actin ± SD.
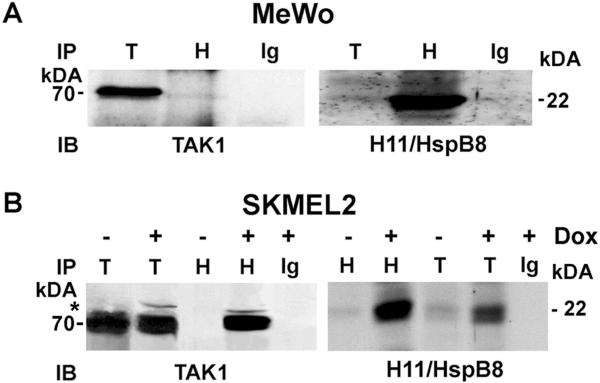
Aza-C resistance is associated with H11/HspB8 mutation
In a last series of experiments, we asked whether H11/HspB8 mutation is associated with resistance to Aza-C treatment. We focused on early passage cultures EK and NL and melanoma cell lines MeWo and HT144, all of which are negative for H11/HspB8 methylation and resist Aza-C-induced cell death. A point mutation (P173H) was seen in MeWo cells, but not in the other cultures. We conclude that Aza-C resistance is associated with this mutation, because H11/HspB8-induced melanoma cell death requires TAK1 binding[23] and P173H did not bind TAK1 in reciprocal pull down assays with H11/HspB8 and TAK1 antibodies (Fig. 9A). By contrast, binding was seen for the wild type protein expressed in SKMEL2 cells (Fig. 9B) that causes cell death when induced by Dox-treatment of cells stably transfected with tet-inducible H11/HspB8[23].
Fig. 9. The H11/HspB8 mutant P173H does not bind TAK1.
(A) Reciprocal pull-down assay of protein extracts from MeWo cells (express P173H) using antibodies to TAK1 (T), H11/HspB8 (H), or normal IgG (Ig) in immunoprecipitation (IP) and immunoblotting (IB). (B) Reciprocal pull-down assay of protein extracts from SKMEL-2 cells stably transfected with tet-regulated H11/HspB8 untreated or treated with Dox (5μg/ml; 3d) done as in (A). Molecular weights are shown on the right. * indicates slower migrating phosphorylated TAK1 protein.
DISCUSSION
The salient features of the data presented in this report are the findings that H11/HspB8 is a growth regulator in normal melanocytes that is silenced by DNA hypermethylation in a high proportion of melanoma tissues and is a potential molecular and therapeutic marker for Aza-C induced melanoma cell death. The following comments seem pertinent with respect to these findings.
Recent interest has focused on DNA methylation as a potential therapeutic target. Methylation of DNA at CpG islands in gene promoters is a relatively common alteration in cancer cells, including melanoma[7,42], and it silences genes involved in all the key pathways that contribute to tumor development and progression, including cell cycle regulation and signalling, differentiation, DNA repair, apoptosis, invasion and immune recognition[43.]. The most frequently mutated gene inherited in familial cutaneous melanoma is CDKN2A that encodes two proteins, p16INK4A and p14ARF, which exert tumor suppressor functions through the pRB and the p53 pathways, respectively. CDKN2A is hypermethylated at loci that are known to independently affect p16INK4A and p14ARF in 27% and 57% of metastatic melanoma samples, respectively[44]. The most frequently hypermethylated genes in cutaneous melanoma are RAR-β2, which mediates growth arrest, differentiation and apoptotic signals triggered by retinoic acids, RASSF1A, which promotes apoptosis and growth arrest, and MGMT, which is involved in DNA repair, being methylated in 70%, 55% and 34% of lesions, respectively[8,45,46]. The list of genes that are hypermethylated in cutaneous melanoma is expanding and it includes new genes (viz. QPCT and LXN) that are hypermethylated in a very high proportion of examined lesions. Re-expression of two of these, HOXB13 and SYK, was shown to reduce colony formation in vitro and diminish tumor formation in vivo, indicating that they function as tumor suppressors in melanoma[10]. Aza-C was also shown to sensitize a melanoma line to interferon-induced apoptosis[46] and DAC upregulated the gene TSPY and suppressed the growth of two derivatives from another line[9]. However, the exact function of most of these genes in melanoma cell death and their ability to serve as markers for the therapeutic efficacy of demethylating agents, if any, remain unknown. Identification of specific biomarkers for individual cancers such as melanoma is particularly relevant as oncology moves forward into the field of personalized therapies and is of crucial importance for patient selection for individual therapies.
We cloned the H11/HspB8 gene from a melanoma cell line and showed that it has an α-crystallin motif and differs from other members of the Hsp family in its biological and functional properties. These include the failure of heat shock to upregulate its expression and cause its nuclear translocation in melanoma cells[14,15,16–22]. Indeed, the presence of an α-crystallin motif does not necessarily imply that a protein functions as an Hsp and a similar motif was seen in the herpes simplex virus type 2 protein ICP10PK that has apoptosis regulatory activity and was used to clone H11/HspB8[21]. In this context it may be important to point out that in one melanoma line, H11/HspB8 carried a single site mutation (W51C) that imparted transforming potential and was able to override the pro-apoptotic activity of the wild type H11/HspB8[20], supporting the interpretation that H11/HspB8 has a central role in melanoma cell life/ death decisions[20,21,23]. H11/HspB8 was also shown to contribute to cell survival and proliferation in skin keratinocytes and cardiomyocytes, apparently involving kinase-dependent Akt activation or inhibition of caspase-3 activation at the level of cytochrome C release[15,18,19, 47,48]. However, we find that in melanocytes, H11/HspB8 causes cell cycle arrest, apparently related to inhibition of the Cyclin E/Cdk2 complex. This inhibition is accompanied by decreased β-catenin phosporylation at Ser552, a site that is involved in the nuclear translocation of β-catenin and the promotion of its transcriptional activity[30,31]. The inhibition of Ser552 phosphorylation is specific, as evidenced by the failure of H11/HspB8 to inhibit phosphorylation of β-catenin at Thr41/Ser45 and Ser675 that are respectively implicated in proteosomal degradation[32,33] or binding of the transcriptional coactivator, CREB binding protein[34]. The mechanism whereby H11/HspB8 specifically inhibits β-catenin phosphorylation at Ser552 is still unclear. We suggest that it involves β-catenin phosphorylation at an interfering site, because: (i) the purified GST-β-catenin protein was phosphorylated by GST-H11/HspB8 but not by GST alone, suggesting that β-catenin is a substrate of H11/HspB8 kinase, and (ii) Ser552 is preceded by a Thr residue (Thr551) the phosphorylation of which could interfere with that of Ser552 as previously shown for adjacent phosphorylation sites in other proteins[36]. However, final conclusions must await the results of ongoing studies to identify the sites of H11/HspB8-mediated β-catenin phosphorylation.
The H11/HspB8 promoter is rich in CpG islands (Supplementary Information, Fig. S1) and we have previously shown that the gene is silenced by DNA hypermethylation in some melanoma cell lines[20,21,23]. However, its methylation in melanoma tissues and the relationship between the levels of methylation, gene expression and melanoma cell death is still unknown. Using MSQP and MSP assays of cell suspensions established from 27 malignant melanoma tissues and 8 early passage melanoma cultures, we found that a high proportion had methylated H11/HspB8 DNA, confirming that methylation is not an artifact unique to established cell lines. The levels of H11/HspB8 methylation differed in various tissues and ranged between 0.06–0.12 units, as also seen for early passage cultures and established cell lines. Overall, the melanoma lines with methylated H11/HspB8 can be classified as having high (> 0.1 units), moderate (0.05–0.1 units) or low (<0.05 units) methylation units that are inversely related to gene expression (mRNA and protein). Cut-off points for the methylated promoter were established from a standard curve after the fluorescence values in the MSQP assays were converted into units of universally methylated DNA, as described by Martinez-Galan et al[26]. Using this calculation, a cut-off point of 0.01 methylation units was established with a sensitivity and specificity of 75–78% for identification of patients with a hypothetical response to demethylation based therapy. We suggest that the levels of H11/HspB8 methylation are a potential marker for the efficacy of demethylating therapy, because: (i) they are significantly related to those of Aza-C-induced melanoma cell death (p<0.001 by Pearson's correlation coefficient), and (ii) high levels of restored H11/HspB8 expression appear to cause robust apoptosis (caspase activation) by overcoming the cell survival functions of the PI3-K/Akt pathway that is known to be constitutively activated in melanoma[37,38]. Indeed, PI3-K inhibition potentiates the ability of demethylation treatment to induce apoptosis in melanoma lines with low or moderate levels of H11/HspB8 methylation. However, HDAC inhibitors did not induce H11/HspB8 expression nor contribute to the death inducing capacity of Aza-C (data not shown), underscoring the specificity of the H11/HspB8 activity.
Importantly, Aza-C caused a significant (p<0.001) decrease in the growth of melanoma xenografts that was also directly related to the levels of H11/HspB8 methylation and restored expression, with high levels being associated with increased inhibition of tumor growth. Similar results were obtained for xenografts established with melanoma cells stably transfected with tet-inducible H11/HspB8 and treated with Dox, confirming the role of H11/HspB8 in demethylation-induced cell death (unpublished observation). Additional support for this conclusion comes from the finding that the P173H mutant expressed by the resistant MeWo cells is not methylated and it does not bind TAK1, an interaction that is required H11/HspB8-induced apoptosis[23]. This is consistent with previous reports for another H11/HspB8 mutant, known as W51C, that was seen in G361 cells and has malignant transformation potential[20,23]. The frequency of H1/HpB8 mutation is still unknown, as only a total of 7 lines were studied to-date. However, given our finding that most studied melanoma cultures are sensitive to Aza-C-mediated cell killing, it is likely that such mutations are relatively rare. Of note, our and previous findings for H11/HspB8 mimic those described for canonical tumor suppressor genes[49–54]. They include the dysfunctional state of H11/HspB8 in melanoma and the fact that its forced expression causes apoptotic cell death. Also characteristic of tumor suppressors is the finding that H11/HspB8 induces cell-type specific growth arrest or survival/proliferation, as reported for keratinocytes and cardiomyocytes[15,18,19, 47, 48]
The EORTC early clinical trials cooperative group experience with Aza-C treatment in patients with metastatic melanoma showed minimal success[55]. In this trial, relatively high doses of Aza-C were given every 5 weeks, and it has been suggested that treatment protocols that administer low doses of Aza-C more frequently over longer periods of time may be more effective because there is a delay in the unfolding of the drug's full demethylating effects[56]. Another consideration is that target genes can become remethylated in the absence of the demethylating agen, such that Aza-C may actually be most effective when given in combination with other drugs. Consistent with this interpretation, we found that PI3-K inhibition enhanced Aza-C-induced cell death. However, HDAC inhibitors had no effect on H11/HspB8 expression or melanoma cell death, whether alone or in combination with Aza-C. Synergy was reported for IFN-β in combination with Aza-C in a melanoma xenograft model[57], and Aza-C was shown to sensitize ovarian cancer xenografts to cisplatin and temolozolmoide, which are two drugs frequently used to treat melanoma. Additionally, Aza-C was shown to enhance the therapeutic activity of IL-2 for melanoma in a phase I clinical trial[58], suggesting that future testing of Aza-C, in combination with other agents is warranted for the treatment of melanoma. However, the identification of molecular and prognostic markers remains a crucial aspect of patient selection for individual targeted treatments. Our data suggest that H11/HspB8 is such a marker for Aza-C treatment of melanoma, whether alone or in combination therapy. Still, given the relatively high numbers of hypermethylated genes and the differential global methylation patterns evidenced by distinct melanoma lines[8–13], it seems reasonable to suggest that panels of hypermethylated genes that include, but are not restricted to H11/HspB8, are likely to provide the widest net for patient selection for demethylating therapy. Our data clearly suggest that H11/HspB8 is an important component of such panels, with patients whose tumors have high levels of H11/HspB8 DNA methylation being seriously considered for demethylation therapy. However, hypermethylation alone is not a sufficient criterion for selecting additional panel members, which also requires a better understanding of their ability to cause melanoma cell death upon restored expression. In the absence of such information, the specific panel make-up is difficult to predict. Ongoing studies are designed to address this question and further verify the therapeutic potential of Aza-C given by other routes and in combination therapy using higher numbers of melanoma samples.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. G. Elias, J. W. Burnett and J. Sinkovics for the gift of the melanoma cell suspensions and cultures.
DECLARATION OF INTEREST This research was supported by Public health Service grant AR053512 from NIAMS, National Institutes of Health (NIH).
REFERENCES
- 1.Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br. J. Dermatol. 2004;150:179–185. doi: 10.1111/j.1365-2133.2004.05708.x. [DOI] [PubMed] [Google Scholar]
- 2.Gershenwald JE, Soong SJ, Balch CM. On behalf of the American Joint Committee on Cancer (AJCC) Melanoma Staging Committee. 2010 TNM Staging. System for Cutaneous Melanoma and Beyond Ann. Surg. Oncol. 2010 Mar 19; Epub ahead of print. [Google Scholar]
- 3.Ghosh P, Chin L. Genetics and genomics of melanoma. Expert Rev. Dermatol. 2009;4:131–143. doi: 10.1586/edm.09.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmieri G, Capone M, Ascierto ML, Gentilcore G, Stroncek DF, Casula M, Sini MC, Palla M, Mozzillo N, Ascierto PA. Main roads to melanoma. J. Transl. Med. 2009;7:86. doi: 10.1186/1479-5876-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O'Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumor specific patterns. Nature Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA, Laird PW. Cancer epigenetics comes of age. Nature Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 7.Howell P, Liu S, Ren S, Behlen C, Fodstad O, Riker AI. Epigenetics in human melanoma. Cancer Control. 2009;16:200–218. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- 8.Furuta J, Umebayashi Y, Miyamato K, Kikuchi K, Otsuka F, Sugimura T, Ushijima T. Promoter methylation profiling of 30 genes in human malignant melanoma. Cancer Sci. 2004;95:962–968. doi: 10.1111/j.1349-7006.2004.tb03184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher WM, Bergin OE, Rafferty M, Kelly ZD, Nolan IM, Fox EJ, Culhane AC, McArdle L, Fraga MF, Hughes L, Currid CA, O'Mahony F, Byrne A, Murphy AA, Moss C, McDonnell S, Stallings RL, Plumb JA, Esteller M, Brown R, Dervan PA, Easty DJ. Multiple markers for melanoma progression regulated by DNA methylation: insights from transcriptomic studies. Carcinogenesis. 2005;26:1856–1867. doi: 10.1093/carcin/bgi152. [DOI] [PubMed] [Google Scholar]
- 10.Muthusamy V, Duraisamy S, Bradbury CM, Hobbs C, Curley DP, Nelson B, Bosenberg M. Epigenetic silencing of novel tumor suppressors in malignant melanoma. Cancer Res. 2006;66:11187–11193. doi: 10.1158/0008-5472.CAN-06-1274. [DOI] [PubMed] [Google Scholar]
- 11.Rothhammer T, Bosserhoff AK. Epigenetic events in malignant melanoma. Pigment Cell Res. 2007;20:92–111. doi: 10.1111/j.1600-0749.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu S, Ren S, Howell P, Fodstad O, Riker AI. Identification of novel epigenetically modified genes in human melanoma via promoter methylation gene profiling. Pigment Cell Melanoma Res. 2008;21:545–558. doi: 10.1111/j.1755-148X.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- 13.Halaban R, Krauthammer M, Pelizzola M, Cheng E, Kovacs D, Sznol M, Ariyan S, Narayan D, Bacchiocchi A, Molinaro A, Kluger Y, Deng M, Tran N, Zhang W, Picardo M, Enghild JJ. Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: clinical implications. PLoS One. 2009;4:e456. doi: 10.1371/journal.pone.0004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CC, Yu YX, Kulka M, Aurelian L. A novel human gene similar to the protein kinase (PK) coding domain of the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10) codes for a serine-threonine PK and is expressed in melanoma cells. J. Biol. Chem. 2000;275:25690–25699. doi: 10.1074/jbc.M002140200. [DOI] [PubMed] [Google Scholar]
- 15.Aurelian L, Smith CC, Winchurch R, Kulka M, Gyotoku T, Zaccaro L, Chrest FJ, Burnett JW. A novel gene expressed in human keratinocytes with long-term in vitro growth potential is required for cell growth. J. Invest. Dermatol. 2001;116:286–295. doi: 10.1046/j.1523-1747.2001.00191.x. [DOI] [PubMed] [Google Scholar]
- 16.Yu X, Heller A, Liehr T, Smith CC, Aurelian L. Expression analysis and chromosome location of a novel gene (H11) associated with the growth of human melanoma cells. Int. J. Oncol. 2001;18:905–911. doi: 10.3892/ijo.18.5.905. [DOI] [PubMed] [Google Scholar]
- 17.Depre C, Hase M, Gaussin V, Zajac A, Wang L, Hittinger L, Ghaleh B, Yu X, Kudej RK, Wagner T, Sadoshima J, Vatner SF. H11 kinase is a novel mediator of cardiac hypertrophy in vivo. Circ. Res. 2002;91:1007–1014. doi: 10.1161/01.res.0000044380.54893.4b. [DOI] [PubMed] [Google Scholar]
- 18.Depre C, Wang L, Sui X, Qiu H, Hong C, Hedhli N, Ginion A, Shah A, Pelat M, Bertrand L, Wagner T, Gaussin V, Vatner SF. H11 kinase prevents myocardial infarction by preemptive preconditioning of the heart. Circ. Res. 2006;98:280–288. doi: 10.1161/01.RES.0000201284.45482.e8. [DOI] [PubMed] [Google Scholar]
- 19.Danan IJ, Rashed ER, Depre C. Therapeutic potential of H11 kinase for the ischemic heart. Cardiovasc. Drug Rev. 2007;25:14–29. doi: 10.1111/j.1527-3466.2007.00002.x. [DOI] [PubMed] [Google Scholar]
- 20.Gober MD, Smith C.C.; Ueda K, Toretsky JA, Aurelian L. Forced expression of the H11 heat shock protein can be regulated by DNA methylation and trigger apoptosis in human cells. J. Biol. Chem. 2003;278:37600–37609. doi: 10.1074/jbc.M303834200. [DOI] [PubMed] [Google Scholar]
- 21.Gober MD, Wales SQ, Aurelian L. Herpes simplex virus type 2 encodes a heat shock protein homologue with apoptosis regulatory functions. Frontiers Biosci. 2005;10:2788–2803. doi: 10.2741/1736. [DOI] [PubMed] [Google Scholar]
- 22.Chowdary TK, Ramon B, Ramakrishna T, Raos CM. Mammalian Hsp22 is a heat inducible small heat-shock protein with chaperone-like activity. Biochem. J. 2004;381:379–387. doi: 10.1042/BJ20031958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li B, Smith CC, Laing JM, Liu L, Aurelian L. Overload of the heat-shock protein H11/HspB8 triggers melanoma cell apoptosis through activation of transforming growth factor-b-activated kinase 1. Oncogene. 2007;26:3521–3531. doi: 10.1038/sj.onc.1210145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danam RP, Howell SR, Remack JS, Brent TP. Heterogeneous methylation of the O(6)-methylguanine-DNA methyltransferase promoter in immortalized IMR90 cell lines. Int J Oncol. 2001;18:1187–1193. doi: 10.3892/ijo.18.6.1187. [DOI] [PubMed] [Google Scholar]
- 25.Ohsie SJ, Sarantopoulos GP, Cochran AJ, Binder SW. Immunohistochemical characteristics of melanoma. J Cutan Pathol. 2008;35:433–444. doi: 10.1111/j.1600-0560.2007.00891.x. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Galan J, Torres B, Del Moral R, Munoz-Gamez JA, Martin-Oliva D, Villalobos M, Nunez MI, Luna Jde D, Oliver FJ, Ruiz de Almodovar JM. Quantitative detection of methylated ESR1 and 14-3-3-sigma gene promoters in serum as candidate biomarkers for diagnosis of breast cancer and evaluation of treatment efficacy. Cancer Biol. Ther. 2008;7:958–965. doi: 10.4161/cbt.7.6.5966. [DOI] [PubMed] [Google Scholar]
- 27.Hatterman K, Mehdorn HM, Mentlein R, Schultka S, Held-Feindt JA. Methylation-specific and SYBR-green-based quantitative polymerase chain reaction technique for O6-methylguanine DNA methyltransferase promoter methylation analysis. Anal. Biochem. 2008;377:62–71. doi: 10.1016/j.ab.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 1995;25:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Resnitzky D, Reed S. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol. Cell. Biol. 1995;27:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He KC, Yin T, Grindley JC, Tian Q, Sato T, Tao WA, Dirsina R, Porter-Wastphal T, Hembree M, Johnson T, Wiedemann LM, Barrett TA, Hood L, Wu H, Li L. PTEN-deficient stem cells initiate intestinal polyposis. Nature Genetics. 2007;39:189–198. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou F, Zhang L, Gong K, Lu G, Sheng B, Wang A, Zhao N, Zhang X, Gong Y. LEF-1 activates the transcription of E2F1. Biochem. Biophys. Res. Comm. 2008;365:149–153. doi: 10.1016/j.bbrc.2007.10.138. [DOI] [PubMed] [Google Scholar]
- 32.Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, Ben-Neriah Y, Alkalay I. Axin-mediated CK1 phosphorylation of beta-catenin at Ser 45: a molecular switch for the WNT pathway. Genes Dev. 2002;16:1066–1076. doi: 10.1101/gad.230302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park CS, Kim SI, Lee MS, Youn CY, Kim DJ, Jho EH, Song WK. Modulation of beta-catenin phosphorylation/degradation by cyclin-dependent kinase 2. J. Biol. Chem. 2004;279:19592–19599. doi: 10.1074/jbc.M314208200. [DOI] [PubMed] [Google Scholar]
- 34.Tauren S, Sanbo N, Qin Y, Browining D, Dulin NO. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J. Biol. Chem. 2006;281:9971–9976. doi: 10.1074/jbc.M508778200. [DOI] [PubMed] [Google Scholar]
- 35.Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mac A, Carpenter M, Smillie LB, Wang JH. Phosphorylation of caldesmon by p34cdc2 kinase. J. Biol. Chem. 1991;266:19971–19975. [PubMed] [Google Scholar]
- 37.Meier F, Schittek B, Busch S, Garbe C, Smalley K, Satyamoorthy K, Li G, Herlyn M. The RAS/RAF/MEK/ERK and PI3K/AKT signaling pathways present molecular targets for the effective treatment of advanced melanoma. Front. Biosci. 2005;10:2986–3001. doi: 10.2741/1755. [DOI] [PubMed] [Google Scholar]
- 38.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat. Rev. Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 39.Dhawan P, Singh AB, Ellis DL, Richmond A. Constitutive activation of Akt/protein kinase B in melanoma leads to up-regulation of nuclear factor-kappaB and tumor progression. Cancer Res. 2002;62:7335–7342. [PubMed] [Google Scholar]
- 40.Stahl JM, Sharma A, Cheung M, Zimmerman M, Cheng JQ, Bosenberg MW, Kester M, Sandirasegarane L, Robertson GP. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 2004;64:7002–7010. doi: 10.1158/0008-5472.CAN-04-1399. [DOI] [PubMed] [Google Scholar]
- 41.Sinnberg T, Lasithiotakis K, Niessner H, Schittek B, Flaherty KT, Kulms D, Maczey E, Campos M, Gogel J, Garbe C, Meier F. Inhibition of PI3K-AKT-mTOR signaling sensitizes melanoma cells to cisplatin and temozolomide. J. Invest. Dermatol. 2009;129:1500–1515. doi: 10.1038/jid.2008.379. [DOI] [PubMed] [Google Scholar]
- 42.Schinke C, Mo Y, Yu Y, Amiri K, Sosman J, Greally J, Verma A. Aberrant DNA methylation in malignant melanoma. Melanoma Res. 2010;20:253–265. doi: 10.1097/CMR.0b013e328338a35a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sigalotti L, Covre A, Fratta E, Paris G, Colizzi F, Rizzo A, Danielli R, Nicolay HJ, Coral S, Maio M. Epigenetics of human cutaneous melanoma: setting the stage for new therapeutic strategies. J Transl Med. 2010;8:56. doi: 10.1186/1479-5876-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freedburg DE, Rigas SH, Russak J, Gai W, Kaplow M, Osman I, Turner F, Randerson-Moor JA, Houghton A, Busam K, Timothy BD, Bastian BC, Newton-Bishop JA, Polsky D. Frequent p16-independent inactivation of p14ARF in human melanoma. J Natl Cancer Inst. 2008;100:784–795. doi: 10.1093/jnci/djn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoon DS, Spugnardi M, Kuo C, Huang SK, Morton DL, Taback B. Profiling epigenetic inactivation of tumor suppressor genes in tumors and plasma from cutaneous melanoma patients. Oncogene. 2004;23:4014–4022. doi: 10.1038/sj.onc.1207505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reu FJ, Leaman DW, Maitra RR, Bae SI, Cherkassky L, Fox MW, Rempinski DR, Beaulieu N, MacLeod AR, Borden EC. Expression of RASSF1A, an epigenetically silenced tumor suppressor, overcomes resistance to apoptosis induction by interferons. Cancer Res. 2006;66:2785–2793. doi: 10.1158/0008-5472.CAN-05-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sui X, Li D, Qiu H, Gaussin V, Depre C. Activation of the bone morphogenetic protein receptor by H11kinase/Hsp22 promotes cardiac cell growth and survival. Circ. Res. 2009;104:887–895. doi: 10.1161/CIRCRESAHA.108.192328. [DOI] [PubMed] [Google Scholar]
- 48.Sanbe A, Daicho T, Mizutani R, Endo T, Miyauchi N, Yamauchi J, Tanonaka K, Glabe C, Tanoue A. Protective effect of geranylgeranylacetone via enhancement of HSPB8 induction in desmin-related cardiomyopathy. PLoS One. 2009;4:e5351. doi: 10.1371/journal.pone.0005351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 50.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 51.St. Clair S, Manfredi JJ. The dual specificity phosphatase Cdc25C is a direct target for transcriptional repression by the tumor suppressor p53. Cell Cycle. 2006;5:709–713. doi: 10.4161/cc.5.7.2628. [DOI] [PubMed] [Google Scholar]
- 52.Prives C, Manfredi JJ. The continuing saga of p53--more sleepless nights ahead. Cell. 2005;19:719–721. doi: 10.1016/j.molcel.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 53.Birch JM, Alston RD, McNally RJ, Evans DG, Kelsey AM, Harris M, Eden OB, Varley JM. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene. 2001;20:4621–4628. doi: 10.1038/sj.onc.1204621. [DOI] [PubMed] [Google Scholar]
- 54.Gasco M, Yulug IG, Crook T. TP53 mutations in familial breast cancer: functional aspects. Hum. Mutat. 2003;3:301–306. doi: 10.1002/humu.10173. [DOI] [PubMed] [Google Scholar]
- 55.Abele R, Clavel M, Dodian P, Bruntsch U, Gunderson S, Smythe J, Renard J, van Glabbeke M, Pinedo HM. The EORTC Early Clinical Trials Cooperative Group experience with 5'-aza-2'-deoxycytidine (NSC 127716) in patients with colo-rectal, head and next, renal carcinomas and malignant melanomas. Eur J Cancer Clin Oncol. 1987;23:1921–1924. doi: 10.1016/0277-5379(87)90060-5. [DOI] [PubMed] [Google Scholar]
- 56.Schwabe M, Lubbert M. Epigenetic lesions in malignant melanoma. Curr Pharm Biotechnol. 2007;8:382–387. doi: 10.2174/138920107783018372. [DOI] [PubMed] [Google Scholar]
- 57.Reu FJ, Bae SI, Cherkassky L, Leaman DW, Lindner D, Beaulieu N, MacLeod AR, Borden EC. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA methylation. J Clin Oncol. 2006;24:3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 58.Gollob JA, Schlambi CJ, Peterson BL, Richmond T, Thoreson M, Moran K, Dressman HK, Jelinek J, Issa JP. Phase I trial of sequential low-dose 5-aza-2'-deoxycytidine plus high-dose intravenous bolus interleukin-2 in patients with melanoma or renal cell carcinoma. Clin Cancer Res. 2006;12:4619–4627. doi: 10.1158/1078-0432.CCR-06-0883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.