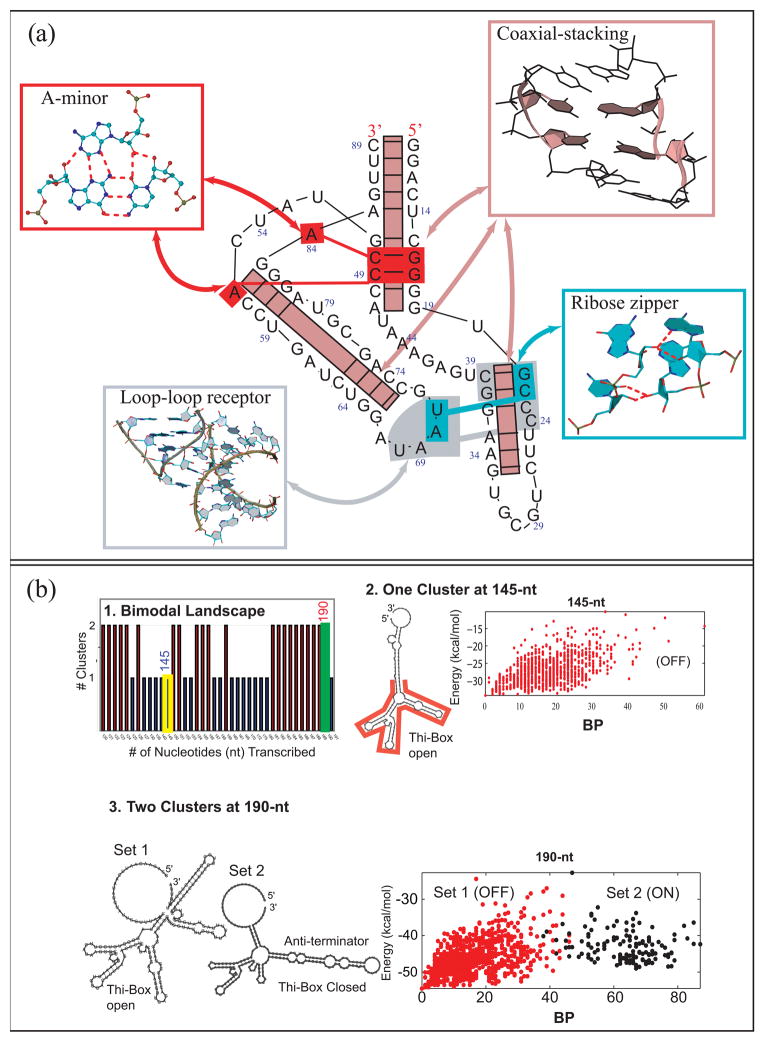
Figure 3.
(a) Annotated diagram of the TPP riboswitch (PDB: 2GDI) shows several correlated motifs working in a cooperative way to stabilize RNA’s 3D conformation. These key motifs can be observed often in many other RNA structures. (b) TPP Riboswitch folding as a function of sequence elongation reveals: (1) either one or two conformational clusters of all suboptimal states for each sequence length. (2) At 145 nt, one cluster is apparent where the folding funnel (base pair difference against free energy plotted for the ensemble for all predicted suboptimal structures) shows a classic simple folding funnel landscape. (3) Near the full sequence length (190 nt), two clusters correspond to the two conformations. See [48] for details.