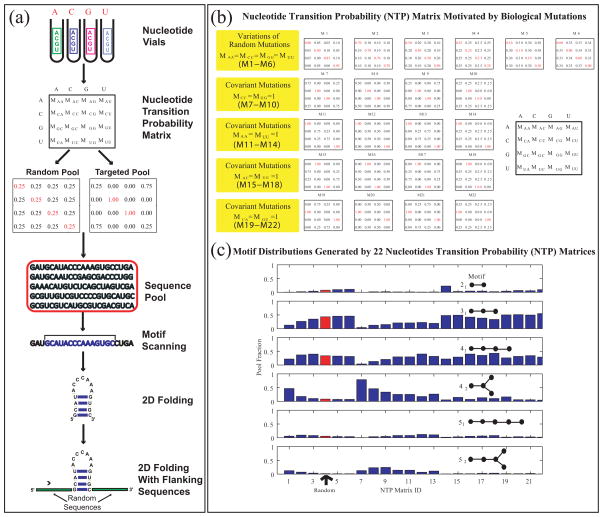
Figure 5.
(a) In Silico Approach to Pool Design. RNA sequence pool generation can be simulated using the nucleotide transition probability matrix, which specifies nucleotide mixtures in the nucleotide vials (or mutation rates for all nucleotide bases). The matrix composition can be defined to be a random matrix, corresponding to experiments, or to mimic specific biological situations, as shown in (b). The resulting sequences can be “folded”' into 2D structures using existing algorithms and analyzed further to screen and filter the candidates against a target motif. The non-random matrices shown in (b) form a basis of 22 probability matrices that generate a wide range of RNA motifs in silico [40], as shown in (c), where motifs yields are organized by RAG graph labels, and the yields of random matrices are shown in red.