Table 6.
Error rates and allele lengths.
| Parameter | Coef | SD(coef) | Amax | 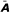 |
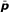 |
|---|---|---|---|---|---|
| GATA417 | 1.59 | 0.38 | 252 | 223 | 0.0512 |
| EV037 | 0.40 | 0.52 | 211 | 203 | 0.0186 |
| GATA028 | 0.40 | 0.52 | 223 | 202 | 0.0186 |
| GT575 | 0.09 | 0.58 | 166 | 155 | 0.0140 |
| GT509 | -0.35 | 0.68 | 217 | 203 | 0.0093 |
| GT310 | -0.35 | 0.68 | 125 | 117 | 0.0093 |
| GT211 | -0.35 | 0.68 | 116 | 106 | 0.0093 |
| GT023 | -0.35 | 0.68 | 115 | 103 | 0.0093 |
| GATA098 | -1.08 | 0.93 | 107 | 93 | 0.0047 |
| EV001 | - | - | 175 | 153 | 0 |
| YEAR | -0.36 | 0.11 | - | - | - |
| Intercept | -4.74 | 0.45 | - | - | - |
| VAR(IND) | 1.81 | 0.94 | - | - | - |
The second column is the values of the coefficients in M1 (Table 2), and the third is their standard deviations. Amax is the length of the largest allele, and 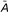 is calculated using the population allele frequencies.
is calculated using the population allele frequencies. 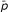 is the mean error rate at the locus. EV001 was not in the analyses, and has therefore no coefficient in M1. Its correct value would have been -∞.
is the mean error rate at the locus. EV001 was not in the analyses, and has therefore no coefficient in M1. Its correct value would have been -∞.
