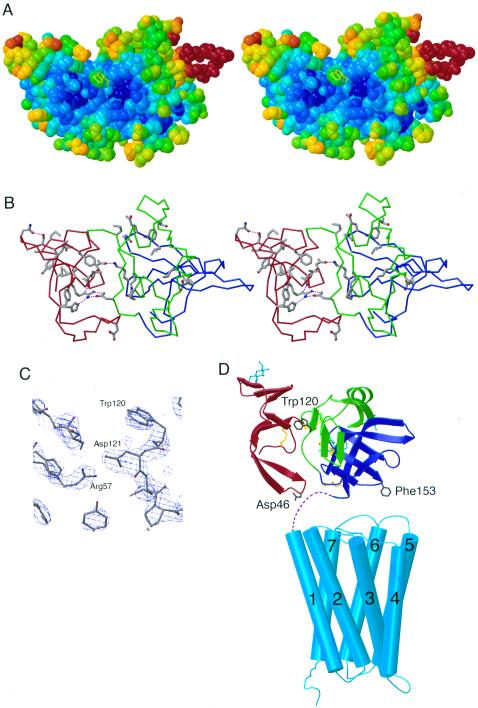
Figure 3.
Possible ligand binding site on Mth. (A) Stereoview of a space-filling model of Mth showing the shallow groove between D1 and D2D3. Atoms are colored according to their temperature factor, with blue and red corresponding to low (relatively well-ordered) and high temperature factors, respectively. Trp-120 (yellow), suggested to be at a ligand binding surface (see text), has a solvent accessible surface area of 126 Å2 [calculated by using a 1.4-Å probe radius with areaimol (26)] compared with 238 Å2 for a fully exposed tryptophan. Examples of protein–protein complexes including critical tryptophan residues at the interface include HFE/transferrin receptor (HFE Trp-81; 115 Å2; second most exposed tryptophan in HFE) (27) and FcRn/Fc (FcRn Trp-133; 136 Å2; third most exposed tryptophan in FcRn) (17). Other receptor-ligand interfaces that include surface-exposed hydrophobic side chains on one of the binding partners are discussed in ref. 17 and references therein. (B) Stereoview of Mth showing residues that are greater than 70% conserved in known Drosophila Mth homologs (excluding conserved cysteines). Asp-121 (conserved in five of the Mth-like proteins and conservatively substituted for glutamate in three others) is involved in an interdomain hydrogen bonds (purple dotted lines) with Arg-57 (conserved in all Mth homologs). (C) Experimental electron density map (MAD solvent-flattened map contoured at 2 σ) in the region near Trp-120. (D) Model for the structure of intact Mth showing relative sizes of Mth ectodomain and the transmembrane region. The Mth transmembrane region is represented by the structure of rhodopsin (21), with adjustments in loop regions to reflect differences in loop lengths between rhodopsin and Mth. Dashed lines represent the seven residues following the Mth ectodomain C terminus and the predicted start of the first transmembrane helix. The Mth ectodomain is oriented such that the C terminus of the model derived from the crystal structure is closest to the membrane. In this orientation, the interdomain groove and Trp-120 are accessible for ligand binding, and two regions containing residues (Asp-46 and Phe-153) suggested to interact with the extracellular face of the seven pass transmembrane domain are positioned near the interhelical loops.