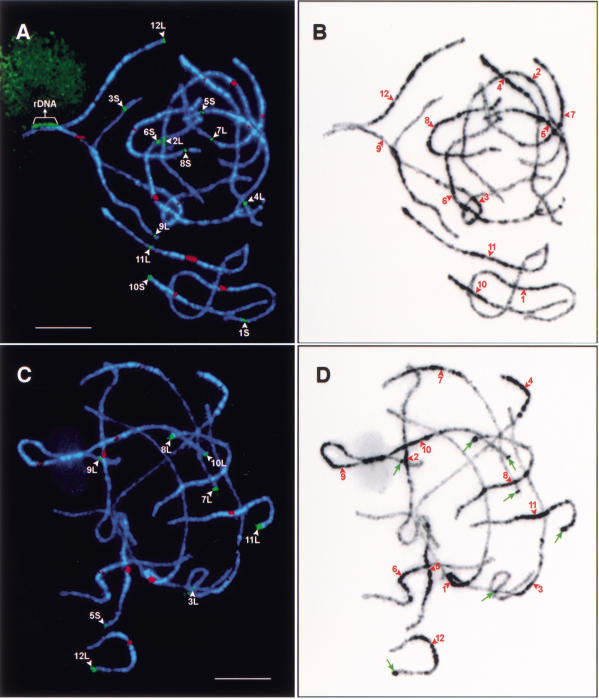
Figure 2.
Karyotyping of rice pachytene chromosomes and mapping of euchromatin and heterochromatin in the rice genome. (A) Chromosomes in a pachytene cell of japonica rice Nipponbare were hybridized with 12 bacterial artificial chromosome (BAC) clones (green signals) that hybridize specifically to each of the 12 rice chromosomes and the centromeric probe pRCS2 (red signals) that hybridizes to all 12 rice centromeres. The 18S·26S rRNA genes are detected on chromosome 9. Bar, 10 μm. (B) DAPI-stained chromosomes in Fig. 2A were converted to a black-and-white image to enhance the visualization of distribution of euchromatin and heterochromatin along the pachytene chromosomes. Note that the centromeres—indicated by red arrowheads—are not always located within a lightly stained chromatin domain. (C) Chromosomes in a pachytene cell of indica rice Zhongxian 3037 were hybridized to the 355-bp tandem repeat Os48 (green signals) and the centromere-specific probe pRCS2 (red signals). Eight Os48 loci (denoted by white arrowheads) were detected and all eight loci, with the exception of the locus on the long arm of chromosome 9, were located on distal regions of the chromosomes. Bar, 5 μm. (D) DAPI-stained chromosomes in Fig. 2C were converted to a black-and-white image to enhance the visualization of distribution of euchromatin and heterochromatin along the pachytene chromosomes. The Os48 loci on chromosomes 3, 7, 8, 9, 10, 11, and 12 are correlated with conspicuous heterochromatin knobs (pointed to by green arrows). The centromeres are indicated by red arrowheads. Note that the heterochromatin distribution patterns in Nipponbare and Zhongxian 3037 are highly similar.