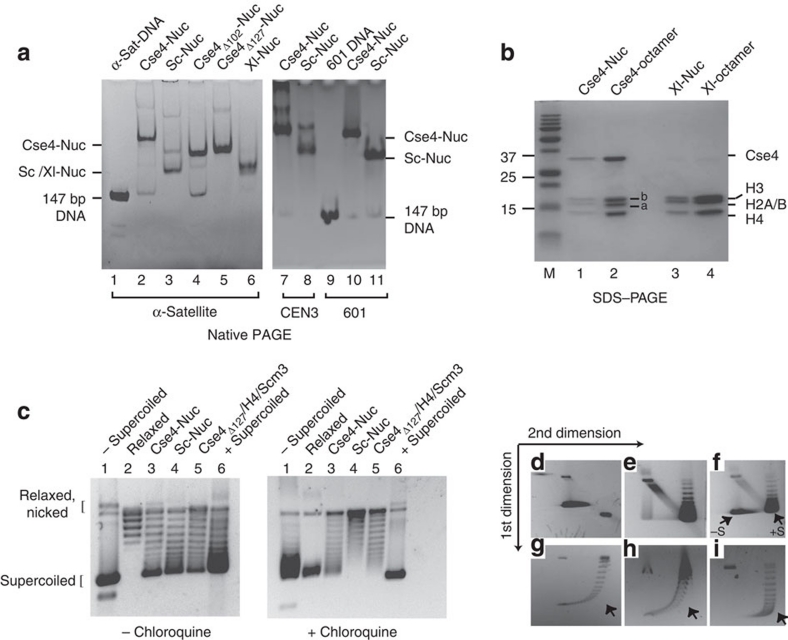
Figure 1. In vitro assembled Cse4-containing nucleosomes exhibit retarded gel mobility and organize DNA in left-handed superhelix.
(a) Cse4-nucleosomes were reconstituted onto 147-bp DNA segments by salt dilution and analysed by 5% native PAGE followed by ethidium bromide staining. Lane 1: 147-bp α-satellite DNA; lanes 2, 4 and 5: nucleosomes with full-length Cse4, Cse4Δ102 and Cse4Δ127, respectively, assembled on α-sat DNA; lanes 3 and 6: S. cerevisiae (Sc) and Xenopus laevis (Xl) nucleosomes on α-sat DNA, respectively. Lanes 7 and 8: Cse4- and Sc-nucleosomes reconstituted on yeast centromeric DNA (CEN3); lane 9: 147-bp 601 DNA; lanes 10 and 11: Cse4- and Xl-nucleosomes reconstituted on 147-bp 601 DNA. (b) Nucleosome bands were eluted from the gel shown in a, lanes 2 and 6, and analysed by SDS–PAGE and Coomassie Blue staining. Lanes 1 and 3: Cse4- and Xl-nucleosome bands; 2 and 4: Cse4- and Xl-octamers; M: protein molecular weight standards. Bands indicated as 'a': yeast H2A; 'b': yeast H2B. (c–i) Cse4-nucleosomes induce negative supercoiling: (c) Nucleosomes were assembled on a circular pBR322 plasmid using yNap1 in the presence of Topoisomerase I. Deproteinized DNA was analysed on 1.2% agarose gel in the absence (left panel) and presence of 2 μg ml−1 chloroquine (right panel). Lane 1: negatively supercoiled plasmid; lane 2: relaxed plasmid; lanes 3 and 4: Cse4- and Sc-nucleosomes, respectively; lane 5: Cse4Δ127/H4/Scm3; lane 6: positively supercoiled pBR322 plasmid (purchased from Inspiralis). (d–i) Two-dimensional gel analysis of plasmid supercoiling. (d) and (e): negatively and positively supercoiled plasmid DNA, respectively, (f): samples in d, e combined; plasmid supercoiling induced by Cse4-nucleosomes (g), Cse4Δ127/H4/Scm3/DNA complex (h) and Sc-nucleosomes (i). −S and +S refer to negatively and positively supercoiled plasmid DNA. The arrow in g–i indicates wherein the most supercoiled topoisomers run if they were positively supercoiled.