Figure 4.
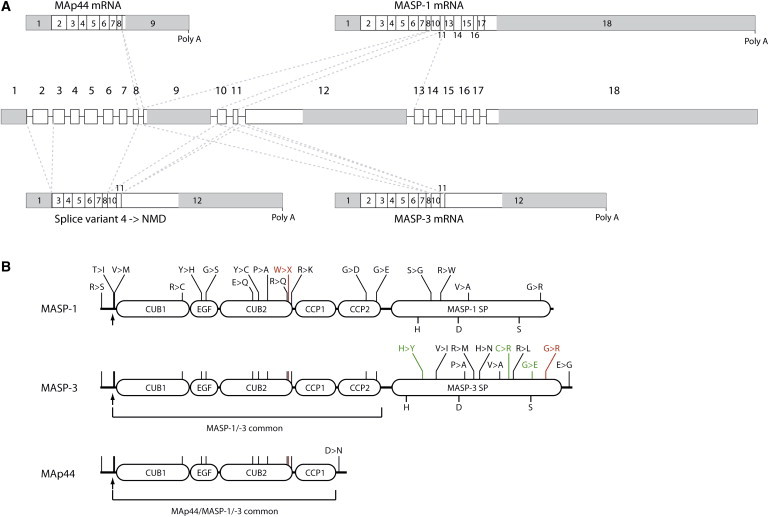
Gene Structure, Alternative Splicing, and Protein Products of MASP1
(A) Overview of the gene or primary transcript and the four resulting alternative splice variants. MAp44, MASP-1, and MASP-3 arise by mutually exclusive differential splicing (indicated by the dotted lines), each product having its own 3′ UTR and poly A signal. A fourth splice product is very similar to MASP-3 mRNA, but with the exclusion of exon 2. This leads to a frameshift, and the product is predicted to be degraded by nonsense-mediated decay.
(B) Overview of the resulting protein structures of the three mRNAs produced from MASP1. The arrow indicates the cleavage site for the signal peptide. Nonsilent polymorphisms are indicated (in black), along with the mutations reported by Sirmaci et al. (in red) and by Rooryck et al. (in green). The exact locations of the indicated nonsilent polymorphisms and mutations are given in Table 2. The part of the polypeptide chain common to all three proteins, as well as the part common to MASP-1 and MASP-3, is indicated.
