Figure 7.
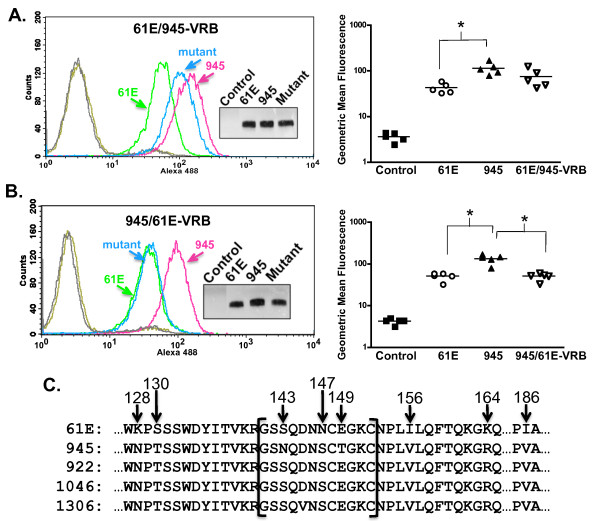
FeLV-945 VRB is sufficient to confer the enhanced binding phenotype to FeLV-A/61E SU. A - B. Comparative flow cytometric binding assays of SU proteins encoded by FeLV-A/61E, FeLV-945 and 61E/945-VRB (in A., left) or the reciprocal mutant, 945/61E-VRB (in B., left). Binding assays were performed using feline 3201 cells as described in Figure 2. Representative histograms are shown, demonstrating the binding activity of FeLV-A/61E SU (green), FeLV-945 SU (pink) and the substitution mutant indicated in each case (blue). Negative controls included supernatants from pCS2/Ctrl-transfected cells (gray) and each mutant SU with isotype control antibody (gold). Inset in each panel shows chemiluminescent western blot analysis to validate equivalent mass amounts of the SU proteins used in the binding assay as previously quantified by infrared dye-based densitometry. Replicate binding assays were performed (right panels) using two (945/61E-VRB), three (61E/945-VRB) or five (FeLV-A/61E and FeLV-945) independently generated and titered batches of SU proteins. The geometric mean fluorescence from individual assays is shown, as is the mean of five independent replicate experiments (horizontal bar). Asterisk indicates statistical significance (*; A: p < 0.01; B: p < 0.001). C. Amino acid sequence of the VRB-containing domain of FeLV/A-61E, compared to that of FeLV-945 and other cohort isolates (FeLV-922, FeLV-1046A, FeLV-1306). Indicated by brackets is consensus VRB, and sequence differences are indicated by the amino acid position number within the mature SU protein.