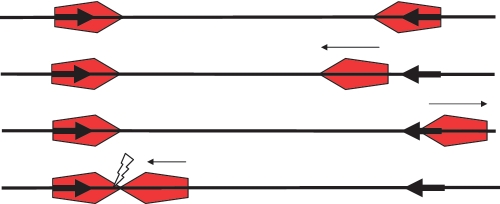
Figure 3.
The MT model on a head-to-head substrate. Small arrows indicate the movement of the enzyme. Top: enzymes (red pentagons with the active NUC domain of the Res subunit located at the leading point of the pentagon) are bound at each target site (bold arrows). Second and third from top: the right-hand enzyme diffuses off its target site after ATP activation and moves randomly backwards and forwards along the DNA. Note that the orientation of the moving enzyme on the DNA is maintained. Low levels of ATP hydrolysis keep the enzyme on the DNA. The left hand enzyme remains at its target. Bottom: the enzyme eventually collides with the left-hand enzyme initiating cleavage (flash). If the ends of the DNA are blocked with an obstacle (such as streptavidin), the moving enzyme generally does not fall off the DNA and cleavage probability is enhanced.