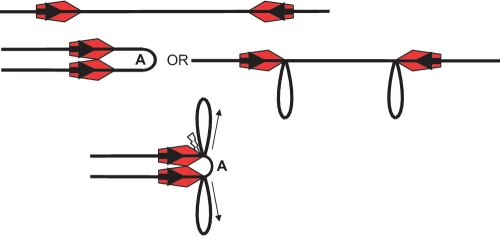
Figure 4.
The AFM model on a head-to-head substrate. Small arrows indicate the movement of the DNA. Top: enzymes (red pentagons with the active NUC domain of the Res subunit located at the leading point of the pentagon) are bound at each target site (bold arrows). Middle: ATP activates both enzyme:enzyme dimerisation (left hand side) and the trapping of loops via the flexibility of the DNA (right hand side). Loop A results from the dimerisation of the enzymes while the other loops result from trapping of DNA segments between the Mod subunit and the Res subunit. By this mechanism the inter-site distance is reduced prior to further translocation. Bottom: translocation, driven by ATP hydrolysis, leading to contraction of loop A in the head-to-head complex, pulls the loop taut between the two translocating Res subunits. It is postulated that this is the trigger for cleavage (flash) by the two NUC domains.