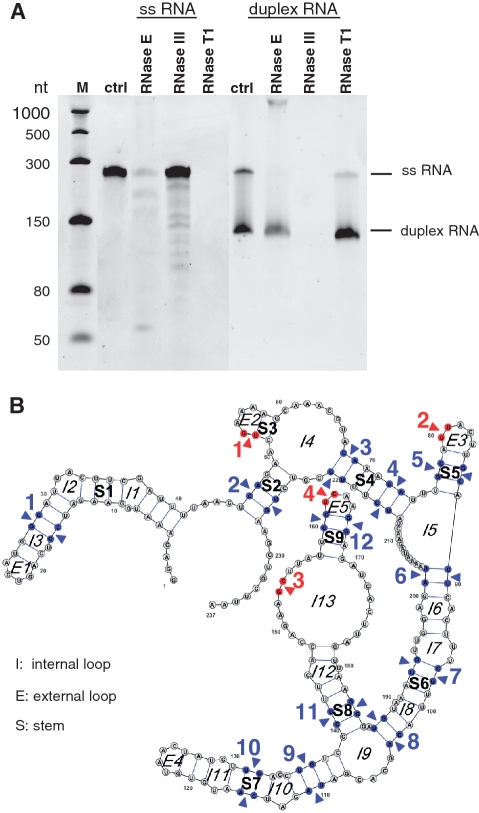
Figure 4.
Ribonuclease cleavage assay of 1.3 pmol single-stranded and 2.6 pmol duplex PMM0685 in vitro RNA. (A) Ethidium bromide-stained 7 M urea–6%PAA gel of PMM0685 single-stranded (ss) and duplex in vitro-synthesized RNA (ctrl) treated with RNase E, RNase III or RNase T1. RNA was incubated with RNases for 15 min. M denotes ssRNA marker from NEB. Though gel electrophoresis was performed under stringent denaturing conditions, only small amounts of duplex RNA were denatured and migrated according to the single-stranded RNA population. (B) The secondary-structure of PMM0685 was folded with RNAfold and drawn using VARNA. RNase E and RNase III cleavage sites, which were deduced from northern blot hybridizations with two oligonucleotide probes targeting the 5′- or 3′-end of PMM0685 in vitro-synthesized RNA, are marked with red arrows or blue arrows, respectively.