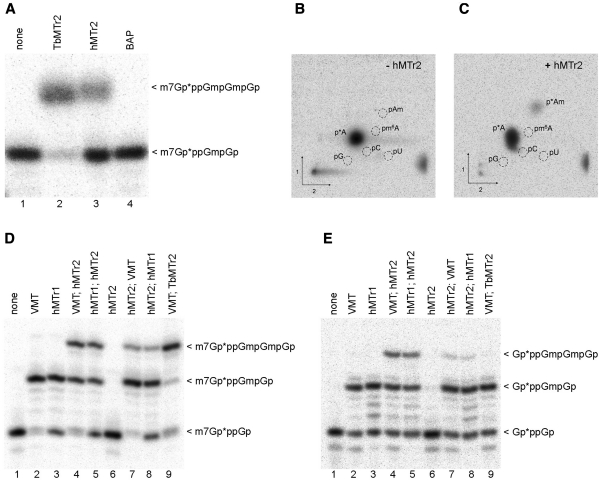
Figure 2.
hMTr2 activity and substrate requirements. Methyltransferase activity: In vitro transcribed RNA-GG molecules with the 32P-labeled cap01 structure (A) were incubated with enzymes as indicated in the presence of SAM. Purified product RNA was digested with RNase T2. Digestion products were resolved on 21% polyacrylamide/8 M urea gel and visualized by autoradiography. BAP protein was used as negative control. RNA with 32P-labeled cap structure created with the TbMTr2 enzyme was used as a reference. Specificity: autoradiography of two-dimensional chromatograms of 5′-phosphate nucleosides on thin layer cellulose plates. [α-32P] ATP-labeled in vitro transcribed cap01-RNA-GA was incubated with SAM in the absence, (B) or presence (C) of the hMTr2 protein. Product RNA was purified, cleaved by nuclease P1 and the resulting nucleotides were analyzed as described (44). 5′-monophosphate ribonucleosides of G, A, U, C, Am and m6A were used as standards. Substrate requirements: In vitro transcribed RNA-GG molecules with 32P-labeled cap0 (D) or capG (E) structure were incubated with one of the indicated enzymes, added in a given order, in the presence of SAM. After every modification step RNA molecules were purified by phenol/chloroform extraction and ethanol precipitation. Final products were digested and analyzed as described in legend for panel A. Asterisks indicate positions of 32P-labeled phosphates.