Abstract
We developed a computer program that can predict the intrinsic promoter activities of primary human DNA sequences. We observed promoter activity using a quantitative luciferase assay and generated a prediction model using multiple linear regression. Our program achieved a prediction accuracy correlation coefficient of 0.87 between the predicted and observed promoter activities. We evaluated the prediction accuracy of the program using massive sequencing analysis of transcriptional start sites in vivo. We found that it is still difficult to predict transcript levels in a strictly quantitative manner in vivo; however, it was possible to select active promoters in a given cell from the other silent promoters. Using this program, we analyzed the transcriptional landscape of the entire human genome. We demonstrate that many human genomic regions have potential promoter activity, and the expression of some previously uncharacterized putatively non-protein-coding transcripts can be explained by our prediction model. Furthermore, we found that nucleosomes occasionally formed open chromatin structures with RNA polymerase II recruitment where the program predicted significant promoter activities, although no transcripts were observed.
INTRODUCTION
It is essential to understand gene regulatory mechanisms to delineate the molecular basis underlying various biological phenomena (1). Particularly intensive efforts have been made to elucidate regulation at the transcription initiation step because it is the first step of gene expression and should play a fundamental role in gene regulation (2–8). Transcription initiation is controlled by an array of cis-regulatory DNA elements to which transcription regulatory proteins, or transcription factors (TFs), bind in a sequence-specific manner (TF binding sites: TFBSs). Subsequently, the bound TFs recruit RNA polymerase II (pol II), and this collectively determines the strength of transcriptional initiation (9,10). It is also supposed that the majority of TFBSs are located in the proximal region of transcriptional start sites (TSSs), i.e. promoters. Therefore, it is hypothesized that analyses of regions upstream of TSSs would elucidate the nature of some of the transcriptional activation activities in the human genome.
After the completion of human genome sequencing (11) and initial gene annotation (12,13), significant efforts have been made to construct a quantitative gene regulatory model based on sequences surrounding TSSs. Using promoter DNA sequence information and expression data, several studies have attempted to explain gene expression levels by examining putative TFBSs in promoter regions (14–24). Various predictive methods have been developed, such as a multiple linear regression model (22), a probabilistic model using Bayesian networks (21), motif expression decomposition (MED) (16,17) and thermodynamic models (14,15). Beer and Tavazoie demonstrated that a Bayesian network model could predict the expression of 2587 yeast genes with an average correlation coefficient of 0.51 by using a subset of 49 clustered microarray expression data sets (21). Nguyen and D'haeseleer applied the MED model and achieved an average correlation coefficient of 0.52 for 5719 yeast genes (17). Gertz et al. (14) predicted the promoter activities of synthetic promoters composed of several known TFBS oligomers by using thermodynamic modeling. Their model predicted promoter activities with a correlation coefficient of 0.66.
Significant progress has been made in predicting gene expression levels, especially when using yeast as a model system (14–17,21). However, the current prediction accuracy is still insufficient, and it remains difficult to apply these previously reported methods to predict promoter activities in human genes. The current difficulty in constructing an accurate model may be caused by the fact that microarray data have been used to monitor expression levels of genes. The microarrays monitor the final levels of gene transcripts. These levels are determined by a number of factors, including the rate of transcriptional initiation and elongation, the efficiency of splicing, the speed of export into the cytoplasm and the rates of degradation (25). Therefore, information from microarray data (and RNA Seq/TSS Seq data, as shown below; also see Supplementary Figure S1) is not a direct indicator of the intrinsic promoter activities of primary DNA sequences. Another drawback to using microarray data is that microarrays essentially monitor relative expression levels and do not represent absolute expression levels.
In our previous article, we reported a systematic luciferase reporter gene assay using HEK293 cells to analyze promoter activities of upstream promoter sequences. These promoter sequences were determined by oligo-capping, which is our full-length cDNA technology (26,27). Using quantitative luciferase assay data to examine promoter activities, we constructed a more accurate quantitative promoter activity prediction model. Additionally, we recently developed TSS Seq, which is a method that combines oligo-capping with massively parallel sequencing (28,29). By TSS Seq analysis, it is possible to massively sequence immediately downstream sequences of TSSs (TSS tags) for analyzing the positions of the TSSs and the frequency of their transcriptions in a given cell type (29,30). Additionally, the digital TSS tag counts can be used as an indicator of absolute expression levels in vivo. We believe that TSS Seq is more suited to our study than original RNA Seq (31), because TSS Seq can simultaneously determine the locations and activities of the transcriptional initiation sites (also see Supplementary Figure S1). In addition, multi-faceted use of the massively paralleled sequencers has provided various types of data, such as the status of the nucleosome structure (micrococcal nuclease-digested genomic DNA sequencing; Nucleosome Seq) and the binding status of RNA polymerase II (pol II; ChIP-Seq) (32–34). We hoped these methods are useful for evaluating the developed prediction model.
In this study, by utilizing luciferase reporter gene data, we constructed a prediction model in which the promoter activity of a given DNA sequence is described as the sum of predicted TFBSs and the transcriptional activation activities of TFs (Figure 1; also see Supplementary Figure S1). We then applied the prediction model to the entire human genome. Comparisons between predicted promoter activities and observed digital TSS tag counts revealed that our prediction model can select active promoters in HEK293 cells from the other silent promoters. Additionally, Nucleosome Seq and pol II ChIP-Seq data revealed that genomic regions with significant prediction scores formed open chromatin structures, and pol II binding was observed, regardless of whether TSS tags were identified from the corresponding genomic region or not. In this article, we describe our first attempt to predict ‘intrinsic’ promoter activities of naked DNA sequences in the human genome.
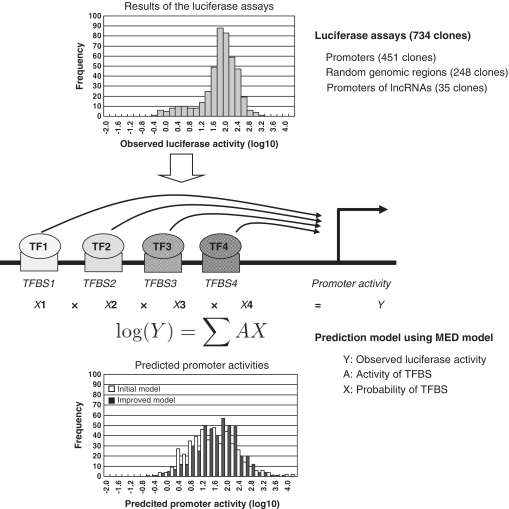
Figure 1.
Schematic representation of the promoter activity prediction model. A schematic of the prediction model is presented here. Distributions of the observed luciferase activities (upper panel) and the predicted promoter activities (lower panel) are also shown.
MATERIALS AND METHODS
Cell culture and luciferase assay
HEK293 cells (ATCC number: CRL-1573) were cultured in DMEM with 10% FBS, kanamycin, 0.15% sodium bicarbonate and 2 mM l-glutamine in 96-well micro-titer plates at a density of 5.0 × 103 cells per well. In each well, 50 ng of promoter clones (451 RefSeq gene promoters, 35 putative lncRNA promoters and 248 random genomic regions) were transiently transfected with 5 ng of pTK-Renilla using 0.3 µl of Fugene 6 (Roche). Forty-eight hours after transfection, dual luciferase assays were performed using a Dual-Luciferase Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions. This procedure was repeated three times with independent cell cultures and transfection experiments. Luciferase activities were divided by Renilla luciferase values and the empty vector pGL3 was used as a plate control. The final transcriptional activities were normalized by the average of the luciferase activities obtained from random genomic regions. The promoter activity data were log-transformed and used to construct the model. Raw luciferase data and sequence information for each of the clones are presented in Supplementary Table S1. See reference (26) for further details.
TSS-Seq, RNA polymerase II ChIP-Seq and Nucleosome Seq analyses
The TSS-Seq library was constructed using HEK293 cells cultured under the same conditions as above, according to the protocols described in Supplementary Figure S2 and reference (28). Our database of TSSs, DBTSS (http://dbtss.hgc.jp/), contains TSS tag information for other cell types (29). RNA pol II ChIP-Seq tag libraries and Nucleosome Seq tag libraries were also constructed from HEK293 cells cultured under the same conditions as described above. Experimental procedures for constructing the ChIP-Seq and Nucleosome Seq libraries are shown in Supplementary Figure S7 and S8. Sequence reads 36 bp long were generated using Illumina GAIIx according to the manufacturer's instructions. The tagged sequences were mapped to the human genome sequence (hg18) using ELAND and no mismatches were allowed. Information on RefSeq genes and putative lncRNAs and other cDNAs are as described in hg18. Statistics of the generated tags are summarized in Table 3. All short read sequences used in this study have been deposited into DDBJ/GenBank under the accession numbers described in Supplementary Figures S2, S7 and S8.
Table 3.
Statistics of sequence tags generated from HEK293 cells and used for validation of the prediction model in vivo
| TSS Seq | No. of total reads | 9 734 314 |
|---|---|---|
| Expected accuracy to detect correct TSSs | 0.9 (also see Supplementary Figure S2C) | |
| No. of total TSS clusters of >5ppm | 6641 | |
| No. of total TSS clusters | 135 579 | |
| Nucleosome Seq | No. of total paired-end reads | 15 071 279 |
| Median insert size | 163 bp | |
| ChIP Seq (pol II) | No. of total reads | 15 864 405 |
| No. of WCE reads | 5 774 736 | |
| No. of IP reads | 10 089 669 | |
| No. of peak detected | 43 214 | |
| No. of peak in RefSeq region (%) | 37 696 (87) | |
| No. of total of TSS Clusters of >5 ppm in HEK293 | 6641 | |
| No. of peak overlapping >5ppm TSS Clusters in HEK293 (%) | 5499 (83) | |
| No. of total of TSS Clusters of <5 ppm in HEK293 | 86 704 | |
| No. of peak overlapping TSS Clusters of <5 ppm in HEK293 (%) | 12 410 (14) |
Prediction model for promoter activities
The prediction model constructed in this study assumed that the promoter activity of a DNA sequence was the sum of the contributions from all TFBS scores using the equation
| (1) |
where Y, A and X represent the observed luciferase activities of the DNA sequence, the number of predicted TFBSs (or the binding probability of the TFBSs) in the DNA sequence and the transcriptional activation score assigned to each TFBS, respectively. Model fitting was conducted using multiple linear regression with the transcriptional activity of a promoter as the dependent variable and the number (or binding probability) of predicted TFBSs as the independent variable.
To search for TFBSs, the TRANSFAC database version 2008.3 was used (35). The parameters to minimize false-positive predictions, as described in TRANSFAC, were used as thresholds for the matrix search conducted by the MATCH algorithm (36). Among the total set of position weight matrices, 192 non-redundant TFBS groups were selected. Twenty-five TFBSs that were identified in less than 4 clones were removed, resulting in a total of 167 TFBSs (see Supplementary Table S2 for the list of TFBSs).
To refine the prediction model, the TRANSFAC matrix score was converted by linear approximation to represent TFBS binding probabilities. The equations describing the binding affinity score are
| (2) |
where x represents the TRANSFAC matrix score, t represents the threshold for the TRANSFAC matrix score and a represents the maximum matrix score. The binding affinity score is assumed to be 0 at the threshold, and it changes linearly above the threshold in 0.1 increments to reach 1.0 at the maximum matrix score. The calculated binding affinity score was used instead of A in the Equation (1) in the gene expression model equation for the improved prediction model. Multiple linear regression models were calculated for each condition and the maximum score giving the best fit was selected. To evaluate the fitting, Pearson's correlation coefficient was calculated between the predicted and observed values of promoter activities. Predicted promoter activities were calculated by leave-one-out cross-validation.
To further improve the prediction model, the search for TFBSs was restricted to the optimum position. DNA sequences were separated into 100-bp bins and the positions considered for TFBSs were extended sequentially from the 3′-end of the DNA. Multiple linear regression models were fitted for each TFBS under each condition, and the position that gave the best fit was selected following a similar procedure as described above.
To select putative TFBSs that had strong effects on transcription, backward stepwise regression based on Akaike's information criterion (AIC) was used.
Validation of the prediction model
To experimentally validate the TFBSs, disruptant mutants were generated and used in luciferase reporter gene assays. Details of plasmids and the results of the luciferase assays are shown in Supplementary Table S4. Experimental procedures for the luciferase assays were as described above.
To evaluate the effects of luciferase gene translational efficiency, a luciferase reporter plasmid containing an internal ribosome entry site (IRES) was constructed as shown in Supplementary Figure S4. DNA fragments were cloned into the IRES luciferase vector system and subjected to luciferase assays. Relative luciferase activities using the IRES vector system were calculated and compared with average luciferase activities observed from cloning random genomic regions into the IRES vector system. Details of the results are presented in Supplementary Figure S4 and Supplementary Table S5.
Previously reported promoter prediction programs
To compare our promoter activity prediction model with previous promoter prediction programs, we used six representative programs: ARTS (37), Eponine (38), EP3 (39), ProSOM (40), Promoter2.0 (41) and FirstEF (42). Programs were downloaded from the following URLs: ARTS scores were downloaded from http://www.fml.tuebingen.mpg.de/raetsch/suppl/arts, ProSOM scores from http://bioinformatics.psb.ugent.be/software/details/ProSOM, Promoter 2.0 scores from http://www.cbs.dtu.dk/cgi-bin/nph-sw_request?promoter and FirstEF scores from the UCSC Genome Browser (http://genome.ucsc.edu/index.html). All programs used promoters or TSSs as inputs. The probability scores produced from these programs were used with the scores from our promoter activity prediction model.
Predicting promoter activity near the 5′-end of human RefSeq genes
RefSeq genes were downloaded from the UCSC Genome Browser (hg18). The promoter regions were defined as the sequence from −1 kb to +200 bp of the 5′-ends of RefSeq genes. To evaluate the ability of our promoter activity prediction model, the Precision and Recall scores were calculated as:
| (3) |
| (4) |
where true positives (TP) are the number of promoter regions with >5 ppm TSS tags and a >1 promoter activity scores; false positives (FP) are the number of promoter regions having no TSS tags and a >1 promoter activity score and false negatives (FN) are the number of promoter regions having >5 ppm TSS tags and a <1 promoter activity score.
To predict potential promoter activities in the human genome, the entire human genome sequence was divided into bins of 1200 bp from the first base of each chromosome. Using the obtained sequences as the input, a promoter activity score was calculated for each bin. The results of predicted promoter activity score for each bin are provided at http://dbtss.hgc.jp/cgi-bin/downloader2.cgi/prediction_score.tar.gz.
RESULTS
Predicting luciferase activities of human primary DNA sequences in HEK293 cells
To construct a model to predict promoter activities of primary human DNA sequences in a given cellular context, we generated a data set of luciferase reporter gene assays using 734 1-kb DNA fragments in HEK293 cells (Figure 1 and Supplementary Table S1). This data set included promoter activity data from 451 DNA regions corresponding to sequences 1 kb upstream of active TSSs. These TSSs were confirmed to be active in this cell line in our previous cDNA sequencing study, namely having 5′-ESTs isolated from HEK293 cells in our oligo-cap cDNA library (3,43). In addition, we collected luciferase data for 248 randomly isolated intergenic DNA fragments (there were no 5′-ESTs in the surrounding regions in any cDNA libraries) and 35 DNA fragments corresponding to sequences upstream of the TSSs of so-called putative intergenic long non-protein coding transcripts (lncRNAs) (44–46), which are also supported by our 5′-oligo-cap ESTs. In total, 83.8% of the promoter clones were from promoters with CpG islands (84.5% were ‘CpG rich’ promoters; see below for evaluation of the model for CpG rich and CpG poor promoters separately).
To predict promoter activities from DNA sequences, we examined putative TFBSs from DNA sequences. We used the TRANSFAC database with parameters to minimize false-positive predictions. Our attempts to optimize the parameters are shown below (35,36). From the total set of TFBS registered in TRANSFAC, we selected and used 167 types of TFBS after removing redundancy among the position weight matrices for the same TFs (see ‘Materials and Methods’ section).
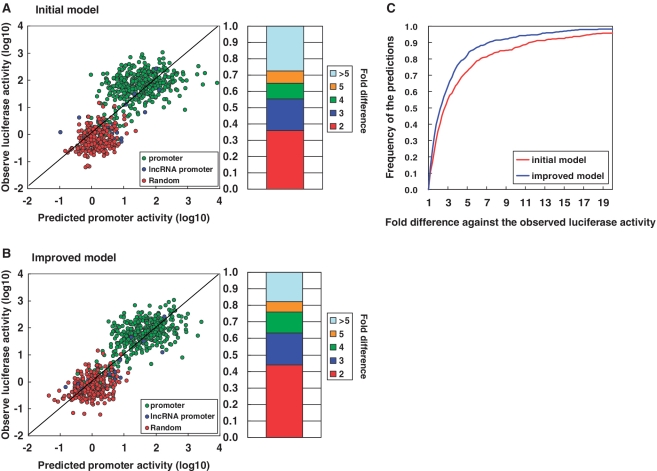
Using the predicted TFBSs and the luciferase activity data, we attempted to explain the luciferase activities with multiple linear regression models (Figure 1). We assumed that log-transformed gene expression levels would be the sum of the contributions from each TFBS (similar to the MED model; see ‘Materials and Methods’ section) (16,17,22). As shown in Figure 2A, the Pearson's correlation coefficient between observed and predicted promoter activities was 0.82. This coefficient was 0.58 when evaluated by only promoter clones and 0.25 when evaluated by only random clones. Although we used the simplest models, which did not consider complex factors such as mutual inter-dependence between individual TFBSs, the accuracy of the constructed prediction model was high. Our model could predict the promoter activity of DNA sequences within a 5-fold range in 73% of the cases (Figure 2A).
Figure 2.
Accuracy of the constructed prediction model. Prediction accuracy of the initial prediction model (A) and the improved prediction model (B). In both (A) and (B), the left panel shows the correlation between the predicted promoter activities (x-axis) and the observed luciferase activities (y-axis). The promoter group and the random genomic sequence group have separate ranges of promoter activities. Because our prediction models have the power to separate these two populations, the overall correlation was 0.82–0.87. Due to this unequal distribution, the prediction accuracy decreased when evaluated separately (0.58–0.66 and 0.25–0.32 for the promoter group and the random genomic sequence group, respectively). The right panel shows the population of predictions for which the difference between the predicted and observed promoter activities are in the range shown in the right margin. (C) The cumulative population of predictions for which the accuracy of the prediction was within the indicated range. Red and blue lines indicate the results of the initial and improved prediction models, respectively.
Fine-tuning the prediction model
We further attempted to improve the promoter activity prediction model by fine-tuning the following parameters: (i) TFBS probability score, we modified the scoring system for predicting TFBSs from DNA sequences (35,36,47) so that the probability of the TFBS was the degree of deviation from the consensus sequence (18,20,48–50), (ii) TFBS position bias to examine positional bias of TFBSs relative to TSSs (51), we introduced different thresholds depending on the relative position of the TFBS to the TSS (Supplementary Table S2) and (iii) feature extraction: we examined the extent to which each TFBS contributed to the accuracy of the prediction model by backward stepwise variable selection using AIC. We found that 85 kinds of TFBS gave the maximum information; thus, these kinds were used as the major determinants of the prediction model. Information about 85 kinds of TFBSs that are present in clone in this study is included in Supplementary Table S3.
Taking these factors together, the prediction model was improved to an eventual Pearson's correlation of 0.87 (0.66 when evaluated only by promoter clones and 0.32 when evaluated by random clones; Figure 2B and C). Contributions from the fine-tuned parameters are summarized in Supplementary Table S2. Details of the computational procedures are also described in the ‘Materials and Methods’ section. The improved version of the prediction model could predict promoter activities within a 5-fold range in 83% of the cases. Information regarding the TFs that made major contributions to the prediction model is shown in Table 1 and also included in Supplementary Table S2.
Table 1.
Promoter activities assigned to the predicted TFBS
| TF ID | TFBS ID | Assigned activity | P-value |
|---|---|---|---|
| Ets1(p54) | V$CETS1P54_02,V$CETS1P54_03 | 0.27 | <2e-16 |
| ZF5 | V$ZF5_B | 0.22 | 1E-12 |
| Myb | V$VMYB_02 | 0.17 | 2E-11 |
| CREB | V$CREB_02,V$CREB_Q4_01 | 0.34 | 3E-11 |
| Sp1 | V$SP1_Q2_01 | 0.30 | 1E-10 |
| ETF | V$ETF_Q6 | 0.21 | 1E-06 |
TFs with P-values of <1 × 106 in stepwise regression validation. Assigned activity scores in the multiple linear regression model are also shown in the third column.
Validation of the prediction model
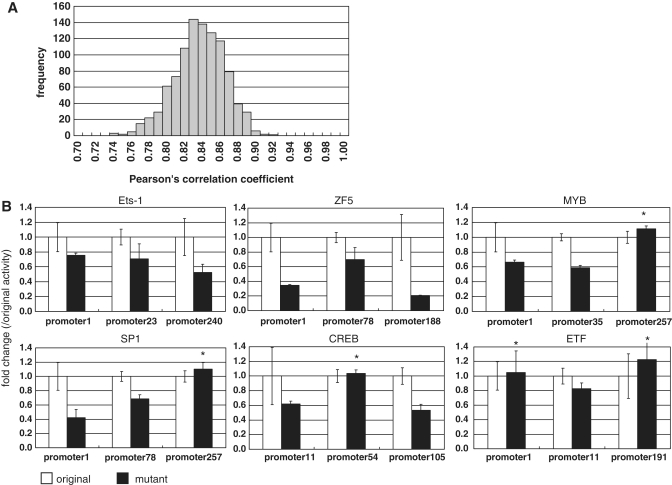
To evaluate the constructed prediction model, we used a 10-fold cross-validation method. With this, we evaluated the risk of over-fitting the regression model. We randomly selected 90% of the luciferase data for the training data set and used the remainder as the test data set. We repeated this test 1000 times and calculated the correlation coefficient for each (Figure 3A). Even in this open test, we found that the average Pearson's correlation coefficient was 0.83 when the fine-tuned prediction model was used, (Figure 3A). These results indicate that the constructed prediction model can be used to predict promoter activities of unknown DNA sequences. This also indicated that over-fitting effects from an excess number of parameters were relatively small in the fine-tuned model.
Figure 3.
Validation of the prediction model. (A) Results of the 10-fold cross-validation test of the model. Distribution of Pearson's correlation between predicted promoter activities and observed luciferase activities is shown. (B) Validation of the TFBSs using promoter disruptant mutants. In each panel, luciferase activities of the original promoter clones (white) and the disruptant mutant clones (gray) are shown. The three upper panels represent the cases in which all promoter clones confirmed the contribution of the predicted TFBS. The three lower panels represent the cases in which predicted changes in luciferase activity were not observed for the examined promoter clones (indicated by the asterisks). In the upper margin in each panel, the ID of the TF corresponding to the examined TFBS is shown. Error bars represent standard deviations calculated from triplicate experiments.
We also examined whether prediction accuracy depended on the base compositions of input DNA or the position weight matrices of the TFBSs. We predicted the promoter activities of input sequences using the following deviated input sequences and position weight matrices: (i) we used randomly generated input sequences with similar average GC content as the known input sequences; (ii) we used promoter sequences from the lower eukaryotes flies, worms and yeast; (iii) we used position weight matrices in which the information order was randomly shuffled. As shown in Supplementary Figure S3, our fine-tuned prediction model could not accurately predict promoter activities when these parameters were altered. Our prediction model uses inherent properties of human genomic sequences and specific mammalian position weight matrices for TFBSs rather than depending on a random combination of sequence information.
To experimentally validate our results, we examined the influence of different translational efficiencies (52) of the luciferase gene on our promoter clones. We evaluated the difference in promoter activities between the usual luciferase vector and a vector where the luciferase gene was translated from an IRES sequence (53). As shown in Supplementary Figure S4, we found that the influence of translational efficacy was very small.
To validate the accuracy of the each of the predicted TFBSs, we evaluated the contributions of the TFBSs to luciferase activities. We constructed promoter clones in which TFBSs were disrupted by site-directed mutagenesis. We compared the promoter activities between the original DNA fragments and the mutagenized DNA fragments. We assayed 24 kinds of TFBSs using 61 mutant DNA fragments. At least 27 (44%) mutants showed significant changes in observed luciferase activities with a false detection rate of P < 0.05 using a t-test (Figure 3B and Supplementary Table S4). These results indicate that at least half of the TFBSs contributing to the prediction model represent truly active TFBSs in HEK293 cells.
Comparison of the prediction mode with previous approaches
We compared the performance of our prediction model with previously reported promoter prediction programs. We tentatively assumed that the prediction score for each promoter prediction reflects its promoter strength. As shown in Table 2, some of the previous promoter prediction programs can be used to predict promoter activities; however, our prediction model gave a higher predictive power than any other program.
Table 2.
Comparison between the performance of our prediction model and previously reported approaches
| This study | ARTS | EP3 | Eponine | ProSOM | Promoter2.0 | FirstEF | ||
|---|---|---|---|---|---|---|---|---|
| a | ||||||||
| All clone | 0.83 | 0.79 | 0.40 | 0.37 | 0.60 | 0.11 | 0.75 | |
| Promoter clone | 0.60 | 0.53 | 0.26 | 0.21 | 0.35 | 0.017 | 0.43 |
| Cell type | ht1080 | g402 | t98g | hct116 | hela | hepg2 | ags | u87mg |
|---|---|---|---|---|---|---|---|---|
| b | ||||||||
| Correlation Coefficient (r) | 0.60 | 0.55 | 0.67 | 0.64 | 0.68 | 0.64 | 0.67 | 0.63 |
aPearson's correlation coefficients between observed luciferase activities and predicted promoter probability scores from the promoter prediction programs are indicated in each column. First line: correlation using the total luciferase data; second line: correlation using luciferase data from the promoter clones only. bPearson's correlation coefficient between luciferase activities and predicted promoter activities from Landolin et al. (54). We constructed the promoter activity prediction model in each cell type independently using the previously published data. Pearson's correlation coefficients were evaluated.
Recently, Landolin et al. (54) reported systematic luciferase assays for 4565 promoters in eight cell types. They described that the activities of ‘ubiquitously’ expressed promoters can be predicted by considering the normalized CG content of the promoters with r = 0.75, although prediction accuracy for the total promoter data set was not specified. They also reported that their predictions became less accurate when high-CG promoters (normalized CG content >0.5) and low-CG promoters (normalized CG content <0.5) were considered separately (r = 0.22 and r = 0.5, respectively). Using our model (r = 0.86 for the total data set and r = 0.66 for promoters only), we evaluated our predictive power similarly. We obtained prediction accuracies of r = 0.34 and r = 0.77 for high- and low-CG promoters, respectively. We also examined whether our models could predict promoter activities using the luciferase data set produced by (54). As shown in Table 2, we constructed a similar prediction model based on luciferase data from the respective cell types. We examined the correlation between the predicted promoter activities and the luciferase data using our constructed models for each cell type, and we found r ≈ 0.6, which was similar to the prediction accuracy we obtained from our original HEK293 data set.
Comparison of the predicted promoter activities with the digital TSS tag counts
We wished to examine the extent to which the prediction model can predict transcriptional activities in vivo. We generated and used a total of 140 million 36-bp TSS tags in HEK293 cells (Table 3). We compared the predicted promoter activities of the region 1 kb upstream of the 5′-ends of RefSeq genes to the digital TSS tag counts observed for the corresponding regions. We observed that the correlation between predicted and observed transcripts was generally low (Supplementary Figure S5), which suggests that it is still difficult to quantitatively predict transcript levels of human genes.
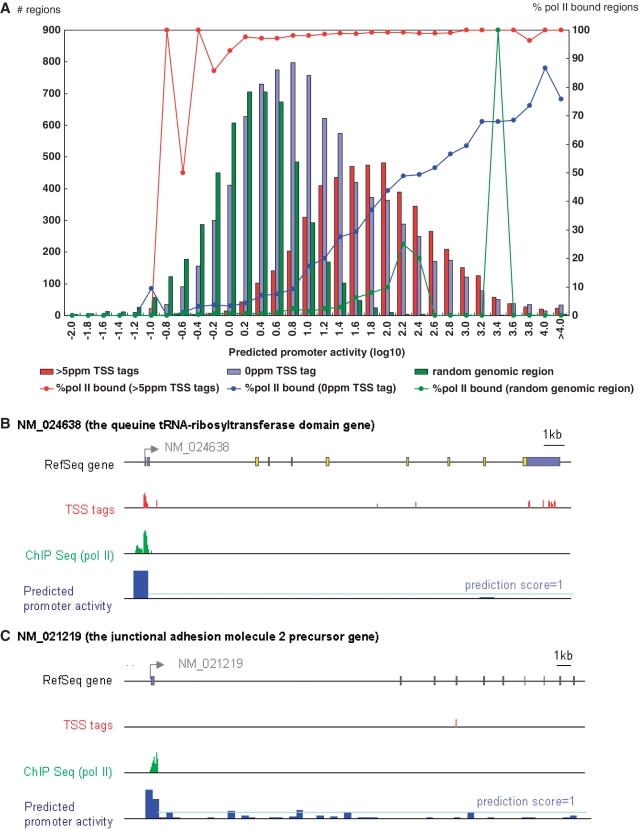
To evaluate the prediction model in a qualitative manner, we examined whether the RefSeq genes with significant expression in HEK293 cells could be separated from silent RefSeq genes. We used a threshold of 5 parts per million (ppm), which is roughly estimated to be five copies of the transcript per cell, assuming that every cell has 1 million mRNA transcripts (28). We determined that promoters with >5 ppm TSS tags should have clearly detectable transcript levels. 5622 cases of RefSeq promoters had 0 < TSS < 5 ppm tag counts and were excluded in this analysis, but the results of a similar analysis using different TSS-Seq tag levels are shown in Supplementary Figure S6.) We compared the distributions of the predicted promoter activities between the RefSeq 5′-end regions with >5 ppm TSS-Seq tags to RefSeq 5′-end regions without TSS tags and to randomly selected genomic regions (Figure 4A). We found clear differences in the distributions between them (P < 1 × 10−100; Wilcoxon rank test). Of 18 686 RefSeq genes, 4749 (25%) had >5 ppm TSS tags in HEK293 cells. Of these, 3922 (83%) had prediction scores >1. Precision and recall of the model to predict TSS tags at this cut-off was (Precision, Recall) = (0.52, 0.83). When TSS tags having >1ppm is also allowed, (Precision, Recall) became (0.63, 0.83). (Precision and Recall using other cut-offs are summarized in Supplementary Figure S6).
Figure 4.
Validation of the prediction model in vivo using TSS-Seq and pol II ChIP-Seq data. (A) Distribution of the 5′-end regions of RefSeq genes with observed TSS tag counts (indicated by the y-axis; left side) and prediction scores (indicated by the x-axis). Red, blue and green bars represent populations indicated in the inset. Red, blue and green lines represent the frequencies of the promoters with pol II binding, as detected by ChIP-Seq (indicated by the y-axis; right side). (B) and (C) are examples of digital TSS tag counts (red bars), pol II binding (green bars) and predicted promoter activities (blue bars) in the RefSeq regions. (B) Exemplifies a case in which all three types of data concordantly indicate the active transcription of the gene. (C) Exemplifies a case in which our model predicted significant promoter activity, although no TSS tags were identified from the corresponding genomic region. The pale-blue line indicates a prediction score of 1.
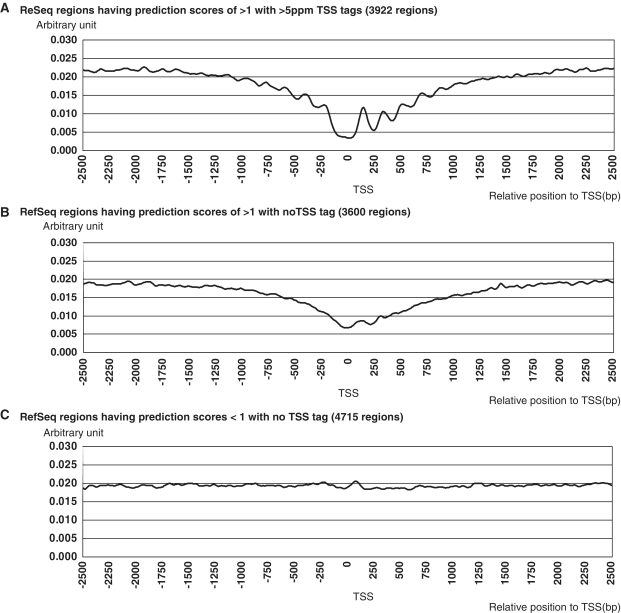
We examined possible causes for the discrepancies between the predictions and the observations. In 3600 cases, the model predicted significant promoter activities with scores of >1, but no TSS tags were observed for the corresponding regions. We generated pol II ChIP-Seq data from HEK293 cells and examined the binding signals of pol II in the surrounding genomic regions for these cases (Table 3; Supplementary Figure S7). Clear binding signals for pol II were observed in 39% of the genomic regions with the prediction scores of >1 and no TSS tags (Figure 4A; also see Figure 4B and C for examples). The pol II binding frequencies increased in proportion to the predicted promoter activities, regardless of whether TSS tags were observed (Figure 4A, blue line). We also examined the nucleosome structure of the surrounding genomic regions. We generated Nucleosome Seq tag data using HEK293 cells and analyzed the nucleosome positioning patterns (Table 3 and Supplementary Figure S8). We found clearly open chromatin structures in genomic regions with prediction scores of >1 and >5 ppm TSS tags (Figure 5A). Interestingly, a similar open chromatin structure was also observed in genomic regions with prediction scores of >1 and no TSS tag (Figure 5B). In these cases, the genomic regions may exhibit significant potential promoter activities, which can be defined as the ability to control the efficacy of forming open chromatin structures and recruiting pol II. Additional factors may inhibit mature formation of the transcripts despite sufficient promoter activity from the upstream DNA sequence. Recent papers have consistently shown that in some cases, pol II rests at the TSS without initiating transcription or transcription is initiated but halts immediately after elongation starts (55–58). It is also possible that transcripts generated from these regions may be aborted during transcription elongation and subjected to rapid RNA degradations, perhaps as polyA minus transcripts. In these cases, our prediction model may have predicted potential promoter activities correctly, though there was discordance with the eventual transcript levels.
Figure 5.
Nucleosome structure at the 5′-end of RefSeq genes. Nucleosome structures are shown for the regions surrounding the 5′-ends of RefSeq genes with having prediction scorers of >1 with >5 ppm TSS tags (3922 regions) (A) having prediction scores of >1 with no TSS tab (3600 regions) (B). Genes having prediction scores <1 with no TSS tab (4715 regions) are shown in (C). For each group, nucleosome occupancy scores (y-axis) were calculated for the indicated genomic position (x-axis) relative to the TSS (or the center of the selected region), according to the method shown in Supplementary Figure S8 and the reference (64). The numbers of regions used for the analyses are shown in the top margins.
Predicted promoter activity landscape of the human genome
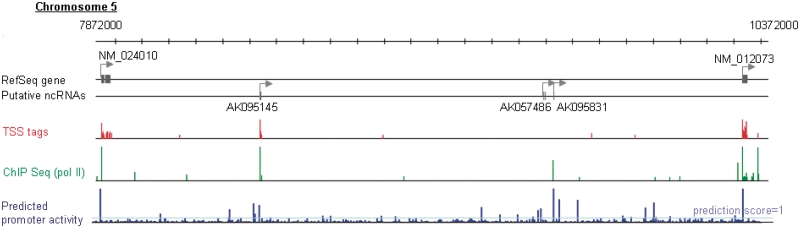
We applied the prediction model to the entire human genome to illustrate the landscape of potential promoter activities in the human genomic sequence. We tentatively defined a prediction score of >1 as the threshold, as used for the RefSeq genes shown above. In total, 185 018 genomic regions outside the RefSeq regions showed prediction scores >1. We examined the overlap between intergenic regions with prediction scores >1 and intergenic regions with >5 ppm TSS-Seq tags. We found 147 overlapping regions. As exemplified in Figure 6, previously identified lncRNA cDNAs were sometimes located in those regions. In these 147 cases, we found that the surrounding genomic regions had an open chromatin structure. Clear binding signals for pol II were observed in 97 cases (66%). These results suggest that biologically controlled transcription is actually occurring from these regions.
Figure 6.
Predicted potential promoter activity landscape of the human genome. Example of an intergenic region with the indicated TSS tag count (red bars), pol II binding signal (green bars) and prediction score (blue bars). The description of this graph is as in Figure 4B and C.
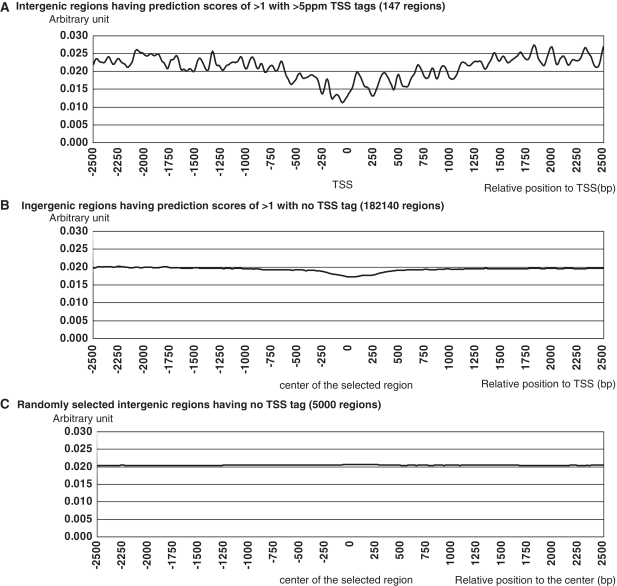
For the remaining 182 140 cases, the genomic regions had prediction scores of >1, but no TSS tags were observed. We examined the nucleosome structure and the binding status of pol II. We found that nucleosome form the open chromatin structure not only in the genomic regions having the prediction scores of >1 with >5ppm TSS tags (Figure 7A) but also in the genomic regions having the prediction scores of >1 without any TSS tags (Figure 7B). These results suggest that many genomic regions had potentially significant promoter activities, although the eventual transcripts from these regions seemed to be repressed by additional factors. Additionally, these results strongly suggest that the current repertoire of intergenic promoter activities or intergenic transcripts, such as lncRNAs, are not derived from experimental errors in cDNA cloning or from randomly occurring and uncontrolled sporadic transcription.
Figure 7.
Nucleosome structure of the intergenic regions. Nucleosome structures are shown for the genomic regions having prediction scores >1 and >5 ppm TSS tags (147 regions) (A), regions having prediction scores >1 with no TSS tags (182 140 regions) (B) and regions having no TSS tags (5000 regions). The description of these graphs is as in Figure 5.
DISCUSSION
In this article, we describe the construction of a model to predict intrinsic promoter activities of primary human DNA sequences. We constructed a reasonably accurate program despite employing a very simple scheme in the prediction model. We believe that the accuracy of the prediction model was achieved by using luciferase data to develop the model. Almost all previous studies have used microarray data for this purpose. In contrast to previous studies, we were able to examine the intrinsic promoter activities of the primary DNA, which may have minimized the effects of other factors. Validation analyses using TSS-Seq, pol II ChIP-Seq and Nucleosome Seq suggested that there are additional factors that significantly contribute to determining the eventual transcript levels, though they seemed to be roughly determined by the promoter activities of the upstream DNA sequences. It should be also noted that utilization of the TSS Seq data, which gives positional information of the TSSs at the same time with their expression levels in a given cellular context in an absolute manner, firstly enabled the evaluation of the constructed prediction model.
In spite of success in predicting promoter activities of primary DNA sequences, we determined that it is still difficult to predict mRNA expression levels in vivo. It is possible that additional factors, as described above, were responsible for the inconsistencies. However, it is also possible that there are inherent problems in massively parallel sequencing analyses. Even TSS-Seq, RNA-Seq, ChIP-Seq and Nucleosome Seq are not strictly quantitative. In some methods, GC-rich sequences are supposed more likely to be represented than AT-rich sequences and in other methods vice versa. Taking such effects into account could possibly further improve the correlation coefficient. Additionally, TSS-Seq and RNA-Seq capture polyA plus RNAs and therefore miss transcripts that are not polyadenylated. The correlation coefficients evaluated by TSS-Seq and RNA-Seq probably underestimate the relevance of predictors that may correctly detect promoters of this class of transcripts. Further validation is necessary for both prediction and evaluation of in vivo transcription events.
An advantage of our model is that TFs and their corresponding TFBSs can be relatively easily identified by analyzing the constructed model. Such separation of factors can sometimes be difficult when more complex models, such as those based on Bayesian networks, are employed. It is unlikely that the transcriptional regulatory network of HEK293 cells consists of only the approximately 200 TFs used for prediction modeling in this study. It is more likely that the TFBSs we used are degenerate to the extent that they coincidentally represent TFBSs for unknown TFs as well. However, even if not all of the TFs that are actually bound to the TFBSs can be identified, our validation using disruptant mutants demonstrated that cis-regulatory elements responsible for transcriptional activation within promoter sequences can be identified in a number of cases (Figure 3B). Also, we were also able to confirm that our model depends on meaningful position weight matrices rather than groups of meaningless complex information units (Supplementary Figure S3C).
We also demonstrated that our approach is valid in different cell types (Table 2). However, further improvements of the model are necessary to predict global patterns of promoter activities in varying cell types. Perturbing the activity score assigned to each TF depending on cell type should be considered for such improvements. Expression information based on digital TSS tag counts of TFs may also be useful to select which TFs are differentially expressed between different cell types. The differences in TF expression may contribute to differential expression of their target transcripts. Indeed, although the prediction model produced by this study is still preliminary, we hope that it will eventually be able to precisely predict the transcriptional landscape of the human genome in a given cellular environment. To this end, sequential improvements of the model should be achieved by considering additional factors, such as distal DNA elements (59), effects of DNA methylation (60,61) and the 3D structure of genomic DNA (62,63).
Such a precise model will be especially useful for interpreting the biological meaning of intergenic lncRNAs, which were identified in large numbers from previous transcriptome studies without any functional inferences. Interestingly, when we applied our prediction model to the entire human genome sequence, we identified tens of thousands of genomic regions that had significant promoter activities potentially without any transcript products. We observed open chromatin structure and clear binding signals of pol II in many cases. Further in-depth experimental validations for detailed analysis of chromatin structure and transcript products from the respective regions are necessary. One step will be to determine whether these are polyadenylated RNAs. It remains unknown whether these genomic regions have any biological relevance or whether they merely represent non-functional promoters that likely occur in a genome as large as that of humans. It is also interesting to examine if these potential promoters, which do not couple with biologically relevant downstream transcripts, can serve as an evolutionary reservoir to construct novel genes or future genomic rearrangements. The biological relevance of the reason why transcription occurs from so many regions throughout the human genome will be first understood by iterative use and improvements of promoter modeling. By presenting the first genome-wide view of the potential promoter activity landscape of the human genome, our prediction model provides a useful starting point toward a comprehensive elucidation of how the code of genomic sequences is decoded into the transcriptome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The New Energy and Industrial Technology Development Organization (NEDO) project of the Ministry of Economy, Trade and Industry (METI) of Japan; the Japan Key Technology Center project of METI of Japan; a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan; a Grant-in-Aid for Scientific Research on Innovative Areas “Genome Science” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT); the Japan Society for the Promotion of Science (JSPS) through its “Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program)”. Funding for open access charge: Grant-in-Aid for Scientific Research on Innovative Areas from MEXT.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to K. Abe, E. Sekimori and K. Imamura for technical support. We are also thankful to Dr Sierro, Dr M. Frith and Mr F. Sathira for critical reading of the article.
REFERENCES
- 1.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki Y, Tsunoda T, Sese J, Taira H, Mizushima-Sugano J, Hata H, Ota T, Isogai T, Tanaka T, Nakamura Y, et al. Identification and characterization of the potential promoter regions of 1031 kinds of human genes. Genome Res. 2001;11:677–684. doi: 10.1101/gr.164001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki Y, Taira H, Tsunoda T, Mizushima-Sugano J, Sese J, Hata H, Ota T, Isogai T, Tanaka T, Morishita S, et al. Diverse transcriptional initiation revealed by fine, large-scale mapping of mRNA start sites. EMBO Rep. 2001;2:388–393. doi: 10.1093/embo-reports/kve085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim TH, Barrera LO, Zheng M, Qu CX, Singer MA, Richmond TA, Wu YN, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto S, Suzuki Y, Kasai Y, Morohoshi K, Yamada T, Sese J, Morishita S, Sugano S, Matsushima K. 5′-end SAGE for the analysis of transcriptional start sites. Nat. Biotechnol. 2004;22:1146–1149. doi: 10.1038/nbt998. [DOI] [PubMed] [Google Scholar]
- 8.Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farnham PJ. Insights from genomic profiling of transcription factors. Nat. Rev. Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Collins FS, Lander ES, Rogers J, Waterston RH, Conso IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 12.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imanishi T, Itoh T, Suzuki Y, O'Donovan C, Fukuchi S, Koyanagi KO, Barrero RA, Tamura T, Yamaguchi-Kabata Y, Tanino M, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gertz J, Cohen BA. Environment-specific combinatorial cis-regulation in synthetic promoters. Mol. Syst. Biol. 2009;5:244. doi: 10.1038/msb.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Zhang J. Accuracy and application of the motif expression decomposition method in dissecting transcriptional regulation. Nucleic Acids Res. 2008;36:3185–3193. doi: 10.1093/nar/gkn127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen DH, D'Haeseleer P. Deciphering principles of transcription regulation in eukaryotic genomes. Mol. Syst. Biol. 2006;2:2006.0012. doi: 10.1038/msb4100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das D, Nahle Z, Zhang MQ. Adaptively inferring human transcriptional subnetworks. Mol. Syst. Biol. 2006;2:2006.0029. doi: 10.1038/msb4100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao F, Foat BC, Bussemaker HJ. Defining transcriptional networks through integrative modeling of mRNA expression and transcription factor binding data. Bmc Bioinformatics. 2004;5:31. doi: 10.1186/1471-2105-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das D, Banerjee N, Zhang MQ. Interacting models of cooperative gene regulation. Proc. Natl Acad. Sci. USA. 2004;101:16234–16239. doi: 10.1073/pnas.0407365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- 22.Bussemaker HJ, Li H, Siggia ED. Regulatory element detection using correlation with expression. Nat. Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Forrest ARR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJL, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EEM. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–U72. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
- 25.Khodursky AB, Bernstein JA. Life after transcription - revisiting the fate of messenger RNA. Trends Genet. 2003;19:113–115. doi: 10.1016/S0168-9525(02)00047-1. [DOI] [PubMed] [Google Scholar]
- 26.Sakakibara Y, Irie T, Suzuki Y, Yamashita R, Wakaguri H, Kanai A, Chiba J, Takagi T, Mizushima-Sugano J, Hashimoto S, et al. Intrinsic promoter activities of primary DNA sequences in the human genome. DNA Res. 2007;14:71–77. doi: 10.1093/dnares/dsm006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki Y, Sugano S. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol. 2003;221:73–91. doi: 10.1385/1-59259-359-3:73. [DOI] [PubMed] [Google Scholar]
- 28.Tsuchihara K, Suzuki Y, Wakaguri H, Irie T, Tanimoto K, Hashimoto S, Matsushima K, Mizushima-Sugano J, Yamashita R, Nakai K, et al. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 2009;37:2249–2263. doi: 10.1093/nar/gkp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita R, Wakaguri H, Sugano S, Suzuki Y, Nakai K. DBTSS provides a tissue specific dynamic view of Transcription Start Sites. Nucleic Acids Res. 2009;38:D98–104. doi: 10.1093/nar/gkp1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 32.Barski A, Cuddapah S, Cui KR, Roh TY, Schones DE, Wang ZB, Wei G, Chepelev I, Zhao KJ. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 33.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang CZ, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 36.Kel AE, Gossling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH (TM): a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonnenburg S, Zien A, Ratsch G. ARTS: accurate recognition of transcription starts in human. Bioinformatics. 2006;22:e472–e480. doi: 10.1093/bioinformatics/btl250. [DOI] [PubMed] [Google Scholar]
- 38.Down TA, Hubbard TJ. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abeel T, Saeys Y, Bonnet E, Rouze P, Van de Peer Y. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Res. 2008;18:310–323. doi: 10.1101/gr.6991408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abeel T, Saeys Y, Rouze P, Van de Peer Y. ProSOM: core promoter prediction based on unsupervised clustering of DNA physical profiles. Bioinformatics. 2008;24:i24–i31. doi: 10.1093/bioinformatics/btn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knudsen S. Promoter2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- 42.Davuluri RV, Grosse I, Zhang MQ. Computational identification of promoters and first exons in the human genome. Nat. Genet. 2001;29:412–417. doi: 10.1038/ng780. [DOI] [PubMed] [Google Scholar]
- 43.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Willingham AT, Gingeras TR. TUF love for “junk” DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bryne JC, Valen E, Tang MHE, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veitia RA. A sigmoidal transcriptional response: cooperativity, synergy and dosage effects. Biol. Rev. 2003;78:149–170. doi: 10.1017/s1464793102006036. [DOI] [PubMed] [Google Scholar]
- 49.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 50.Das D, Pellegrini M, Gray JW. A primer on regression methods for decoding cis-regulatory logic. PLoS Comput. Biol. 2009;5:e1000269. doi: 10.1371/journal.pcbi.1000269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koudritsky M, Domany E. Positional distribution of human transcription factor binding sites. Nucleic Acids Res. 2008;36:6795–6805. doi: 10.1093/nar/gkn752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray NK, Wickens M. Control of translation initiation in animals. Annu. Rev. Cell. Dev. Bi. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 53.Bochkov YA, Palmenberg AC. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. Biotechniques. 2006;41:283. doi: 10.2144/000112243. [DOI] [PubMed] [Google Scholar]
- 54.Landolin JM, Johnson DS, Trinklein ND, Aldred SF, Medina C, Shulha H, Weng ZP, Myers RM. Sequence features that drive human promoter function and tissue specificity. Genome Res. 2010;20:890–898. doi: 10.1101/gr.100370.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- 56.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu JY, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 64.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.