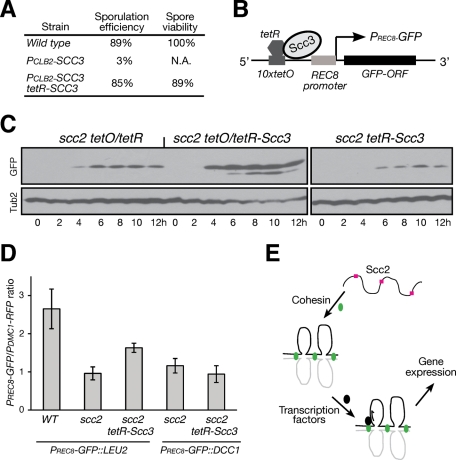
FIGURE 8:
Activation of REC8 promoter by forced localization of cohesin. (A) Generating a functional tetR-SCC3 fusion allele. Yeast strains (NH144, 3200, and HY2636) were sporulated, and tetrads were dissected for determination of spore viability. We used the CLB2 promoter to replace the endogenous SCC3 promoter to generate PCLB2-SCC3. (B) A schematic diagram showing forced localization of Scc3, and therefore cohesin, to the 10× copies of tetO sequence located 5′ upstream of the REC8 promoter. The distance between the first tetO and the REC8 promoter is ∼2 kb. (C) Immunoblot showing the production of GFP. Cells were induced to undergo synchronous meiosis, and protein extracts were prepared for immunoblotting. Strains used: HY2685 (scc2 tetO), HY2684 (scc2 tetO/tetR-Scc3), and HY2692 (scc2 tetR-Scc3). (D) Quantification of PREC8-GFP production. Yeast cells were induced to undergo synchronous meiosis, and the fluorescence intensities of GFP and RFP were determined as for Figure 5C. Strains used: WT, HY3100; scc2 PREC8-GFP::LEU2, HY3044; scc2 tetR-SCC3 PREC8-GFP::LEU2, HY3045; scc2 PREC8-GFP::DCC1, HY3046; and scc2 tetR-SCC3 PREC8-GFP::DCC1, HY3047. All strains harbor one copy of PDMC1-mAPPLE (RFP) and 10 copies of tetO inserted at the LEU2 locus. (E) A model depicts Scc2 and cohesin action in transcriptional activation during meiosis. Scc2, red squares; cohesin, green ovals. Black ovals represent an unknown transcription factor. Chromosomes are shown as black and gray lines.