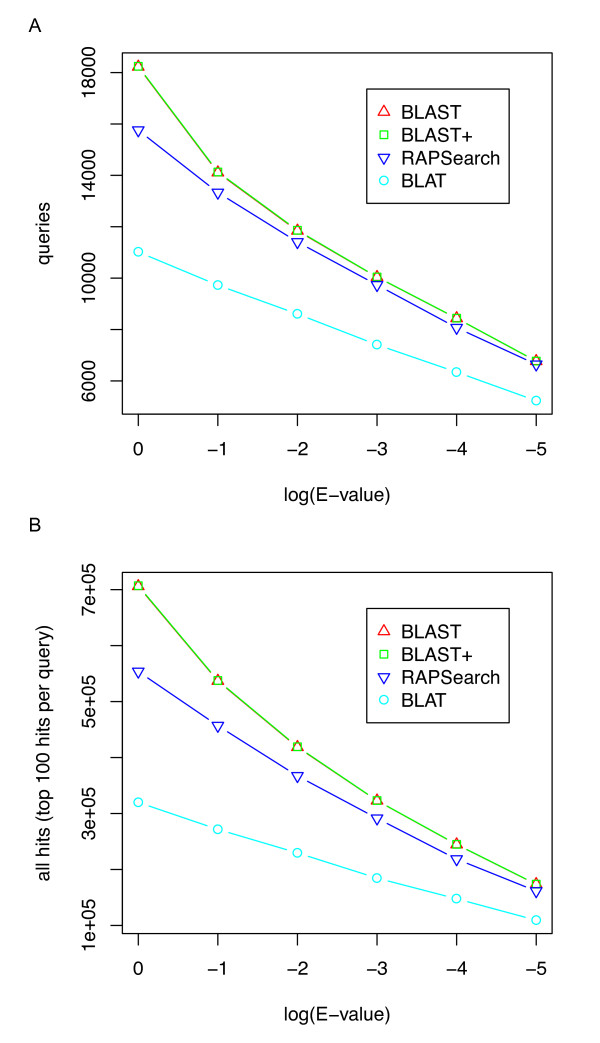
Figure 3.
Comparison of the similarity search sensitivity at different E-value cutoffs. RAPSearch, BLAST (and BLAST+, using default parameters), and (protein) BLAT were compared on the same query dataset (4440037). The total number of queries that have at least one homolog in the IMG protein sequence database (based on the corresponding E-value cutoff) was used in (A), whereas the total number of all significant hits (up to 100 hits per query) was used in (B). Note that BLAST and BLAST+ have almost identical sensitivity (but BLAST+ is twice as slow). For this dataset search, BLAST, RAPSearch and BLAT used 154, 3.5 and 2.7 CPU hours (on Intel Xeon 2.93 GHz), respectively.