Abstract
We previously demonstrated that streptokinase (SK) can be used to generate active site-labeled fluorescent analogs of plasminogen (Pg) by virtue of its non-proteolytic activation of the zymogen. The method is versatile and allows for stoichiometric and active site-specific incorporation of any one of many molecular probes. The limitation of the labeling approach is that it is both time-consuming and low yield. Here we demonstrate an improved method for the preparation of labeled Pg analogs by the use of an engineered SK mutant fusion protein with both COOH- and NH2-terminal His6-tags. The NH2-terminal tag is followed by a tobacco etch virus proteinase cleavage site to ensure that the SK Ile1 residue, essential for conformational activation of Pg, is preserved. The SK COOH-terminal Lys414 residue and residues Arg253-Leu260 in the SK β-domain were deleted to prevent cleavage by plasmin (Pm), and to disable Pg substrate binding to the SK·Pg*/Pm catalytic complexes, respectively. Near-elimination of Pm generation with the SKΔ(R253-L260)ΔK414-His6 mutant increased the yield of labeled Pg 2.6-fold and reduced the time required >2-fold. The versatility of the labeling method was extended to the application of Pg labeled with a near-infrared probe to quantitate Pg receptors on immune cells by flow cytometry.
Keywords: plasminogen, streptokinase, fluorescence probes, binding, kinetics, plasminogen receptors
Introduction
The streptococcal pathogenicity factor and thrombolytic drug, streptokinase (SK)1 activates the human fibrinolytic system by activation of the serine proteinase zymogen, plasminogen (Pg) to the fibrin clot-dissolving proteinase, plasmin (Pm). [Glu]Pg is the circulating form of Pg, which consists of a 77-residue NH2-terminal PAN (Pg/apple/nematode) module [1; 2], five homologous kringle domains, of which K1, K4, and K5 contain high to moderate affinity lysine-binding sites (LBS) [3], and a COOH-terminal serine proteinase zymogen domain [4]. The NH2-terminal PAN module is jettisoned by Pm cleavage of [Glu]Pg to yield [Lys]Pg, which is more readily activated by the SK·Pg* and SK·Pm catalytic complexes [5; 6; 7]. As shown in the crystal structure of SK bound to the isolated catalytic domain of Pm (μPm), SK consists of three β-grasp domains, α, β, and γ, linked by flexible segments [8]. SK has a flexible domain structure in solution, but assumes a well defined three-sided crater-like structure around the catalytic site of μPm [8; 9; 10].
The mechanism of Pg activation by SK is initiated by rapid formation of an SK·Pg* complex in which the zymogen is activated non-proteolytically by the molecular sexuality mechanism [7; 11; 12; 13]. Although there is no crystallographic proof, the Ile1 NH2-terminus of SK inserts into the NH2-terminal binding cleft in the Pg catalytic domain, forming a salt bridge with Asp194 (chymotrypsinogen numbering), which induces conformational activation of the catalytic site. Subsequently, tightly coupled with conformational activation, Pg binds as a specific substrate of the SK·Pg* complex, which is cleaved at Arg561-Val562 to form Pm. Pm binds to SK with ~830-fold higher affinity than [Lys]Pg, resulting in displacement of Pg from the SK·Pg* complex and formation of SK·Pm, which also recognizes Pg as a specific substrate and converts the remaining free Pg to Pm [5; 7; 14].
We previously developed analogs of the Pg zymogen specifically labeled at the active site with fluorescence probes by trapping, and thus inactivating, the transiently formed SK·Pg* complex with a thioester tripeptide chloromethyl ketone. Following deacetylation of the NH2-terminal thioester of the incorporated inhibitor with NH2OH under mild conditions, the unique free thiol can be modified with thiol-reactive fluorescence probes [7; 14; 15; 16]. Development of the fluorescent Pg analogs enabled the reversible SK-Pg binding and conformational activation processes to be quantitated for the first time, uncoupled from proteolytic generation of Pm [7; 14; 15; 16; 17; 18]. The labeling approach is versatile in that any thiol-reactive molecular probe may be used [19]. Peptide chloromethyl ketones require use of the catalytic apparatus to inactivate serine proteinases, and are thus chemically specific for labeling proteinases varying greatly in substrate specificity [20]. The labeling approach has also been used to active site-specifically immobilize Pm and other proteinases on chromatography matrices [21], and for incorporation of heavy atom derivatives for crystallography [22]. For employing fluorescence probes to investigate the mechanism of Pg activation, the labeling scheme has the advantage of allowing labeling of Pg and Pm with any of the hundreds of commercially available, thiol-reactive probes with widely different spectral properties. This is an advantage because generally it is not possible to predict which probe will provide a useful fluorescence signal for quantitation of a particular interaction.
A limitation of the fluorescent Pg analogs is the difficulty of their preparation and relatively low yield. The principal source of the problem is the highly efficient coupling between conformational Pg activation and proteolytic generation of Pm at the expense of Pg [5]. The conformational and proteolytic activation reactions of the mechanism are regulated by kringle-mediated LBS interactions [5; 7; 14; 15; 16; 23; 24; 25; 26]. Binding of the lysine analog, 6-aminohexanoic acid (6-AHA) to a Pg and Pm kringle domain weakens SK·Pg* and SK·Pm catalytic complex formation by blocking binding of the SK COOH-terminal Lys414 residue to a Pg/Pm kringle [27]. Interaction of Pg as a substrate of the catalytic complexes is also greatly inhibited by 6-AHA. We recently identified Arg253, Lys256, and Lys257 in the SK β-domain 250-loop as the residues responsible for facilitating LBS-dependent Pg substrate recognition, mediated by kringle 5 of Pg [28].
The goals of the present studies were to engineer an SK construct in which conformational activation was preserved and LBS interactions involved in Pm generation were diminished to enable improved preparation of active site-labeled Pg analogs. Steady-state kinetic approaches developed previously [5; 7; 27; 28] that separate conformational and proteolytic activation, and equilibrium binding approaches using fluorescent Pg analogs were applied to evaluate the properties of SK mutants designed for this purpose. A double His6-tagged SK was constructed that contains an NH2-terminal His6-tag with a tobacco etch virus proteinase (TEVp) cleavage site abutted to the essential Ile1, which lacks Lys414, contains a COOH-terminal His6-tag, and in which the residues Arg253-Leu260 of the 250-loop in the β-domain were deleted. Following TEVp cleavage of His6-(TEVp cleavage site)-SKΔ(R253-L260)ΔK414-His6 to generate Ile1, the mutant was characterized in kinetic and equilibrium binding studies of [Lys]Pg. SKΔ(R253-L260)ΔK414-His6 retains conformational activation of [Lys]Pg and has greatly reduced activity in proteolytic generation of Pm. Active site-labeling with the new SK construct simplified the procedure, increased the yield of labeled Pg analogs, while maintaining high purity. In a new application of the labeling procedure, [Lys]Pg was labeled with the near-infrared probe, Alexa Fluor 680, and then used for FACS analysis to identify cells rich in Pg receptors from murine blood and spleen.
Materials and methods
Native SK and an SK mutant with NH2- and COOH-terminal His6-tags
Native SK (SK) was prepared as previously described [14; 16]. An SK fusion protein construct was prepared that encoded wtSK flanked on either side by His6-tags. Site-directed mutagenesis was performed using QuikChange site-directed mutagenesis (Stratagene) with 5′- and 3′-PCR primers that eliminated the stop codon that was described previously to be present in the His6-wtSK [27]. The resulting constructs created a wtSK fusion protein flanked by His6-tags (bold) with a TEVp cleavage site (underlined) encoded before the wtSK protein, Met-His6-Ser-Ser-Gly-Leu-Val-Pro-Trp-Asn-Glu-Asn-Leu-Try-Phe-Gln-SKIle1…SKK414-Ile-Arg-Gly-Gly-Ser-Pro-Gly-Leu-Gln-Glu-Phe-Asp-Ile-Lys-Leu-Ile-Asp-Thr-Val-Asp-Leu-Glu-His6. This construct, designated His6-wtSK-His6, has an 8-residue linker between the NH2-terminal His6-tag and the TEVp cleavage site and a 22-residue linker between Lys414 and the COOH-terminal His6-tag, which ensure accessibility of the His6-tags.
Generation, expression, and purification of SKΔ(R253-L260)ΔK414)-His6
Deletion of R253-L260 in the β-domain of wtSK was achieved by site-directed mutagenesis on the His6-wtSK-His6 gene. 5′- and 3′-PCR primers flanking the loop (R253- L260, underlined) were created with the sense primer 5′-CGG GAA CAA GCT TAT AAT GAA GAA ATA AAC AAC ACT GAC CTG-3′ and antisense primer 5′-CAG GTC AGT GTT GTT TAT TTC TTC ATT ATA AGC TTG TTC CCG-3′. Deletion of Lys414 on the His6-SKΔ(R253-L260)-His6 template was made using sense primer 5′-CCT GAT AAC CCT AAC GAC GGC TCC CCC GGG CTG -3′ and antisense primer 5′-CAG CCC GGG GGA TCC GCC GTC GTT AGG GTT ATC AGG-3′. Mutations were confirmed by DNA sequencing. Both His6-wtSK-His6/pET30b(+) and His6-SKΔ(R253-L260)ΔK414-His6/pET30b(+) were transformed in E. coli Rosetta (DE3) pLysS cells for protein expression.
Proteins were expressed by induction with 0.5 mM isopropyl-D-thiogalactopyranoside for 4 h at 37 °C. Cells were harvested by centrifugation, resuspended in 50 mM Hepes, 125 mM NaCl, 1 mg/ml polyethylene glycol (PEG) 8000, pH 7.4 (Buffer A) with 1 mM EDTA and 0.2 % sodium azide, lysed by 3 cycles of freeze-thaw, and centrifuged to clarify the lysates. His6-wtSK-His6 or His6-SKΔ(R253-L260) ΔK414-His6 inclusion bodies were solubilized by addition of Buffer A containing 3 M NaSCN followed by vortexing and centrifugation. The lysate was dialyzed into 50 mM Hepes, 400 mM NaCl, 50 mM imidazole, pH 7.4 (Buffer B) and purified by Ni2+-iminodiacetic acid-Sepharose chromatography with a 50–500 mM imidazole gradient in Buffer B. TEVp-His6 was added to the eluted protein in a 1:10 molar ratio of enzyme to substrate. Both a previous study of the substrate specificity of TEVp [29] and our studies of recombinant staphylocoagulase [30] indicated that TEVp has reduced cleavage efficiency (~50% at saturating substrate) for Ile at the P1′ position3 (Panizzi, P. and Bock, P. E. unpublished observations), compared to Gly, Ser, or Ala at P1′, which are optimal for TEVp cleavage [29]. The reaction mixture was first dialyzed overnight into 50 mM Hepes, 300 mM NaCl, 1 mM DTT, 5% glycerol, pH 7.8 at 4 °C and subsequently dialyzed back into Buffer B. Uncleaved His6-wtSK-His6 or His6-SKΔ(R253-L260) ΔK414-His6, cleaved wtSK-His6 or SKΔ(R253-L260)ΔK414-His6, and TEVp-His6 bound to Ni2+-iminodiacetic acid-Sepharose with different apparent affinities that resulted in two relatively equivalent primary peaks eluted by the shallow 30 column-volume imidazole gradient (50–500 mM). The first peak was wtSK-His6 or SKΔ(R253-L260)ΔK414-His6 and the second peak was His6-wtSK-His6 or His6-SKΔ(R253-L260)ΔK414-His6. wtSK-His6 or SKΔ(R253-L260) ΔK414-His6 was pooled, concentrated, dialyzed against Buffer A without PEG, quick-frozen, and stored at −80°C. NH2-terminal sequencing of the polyvinyldiene fluoride membrane-blotted proteins confirmed the NH2-terminal sequence for wtSK-His6 (Ile-Ala-Gly-Pro-Glu) and for His6-wtSK-His6 (Met-His-His-His-His).
TEV proteinase
A TEVp mutant designed to minimize autolytic proteolysis (Ser219 to Val) and containing a COOH-terminal His6-tag was kindly provided by Dr. Laura Mizoue. TEVp-His6 was expressed in Rosetta (DE3) plysS cells, induced with 1 M isopropyl-D-thiogalactoside at a 600 nm-optical density of ~0.5 for 3 h at 37 °C, and cells were harvested by centrifugation. Recovered cells were resuspended in Buffer B containing 1 mg/ml PEG 8000, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 2 mM DTT (Buffer C). Cells were lysed by sonication followed by incubation with DNAse I (20 μg/ml) and 80 mM MgCl2 for 1 h with constant stirring at 25 °C, and centrifuged. TEVp-His6 was purified from the supernatant by chromatography on Ni2+-iminodiacetic acid-Sepharose by elution with a 50–500 mM imidazole gradient in Buffer C. Fractions containing the proteinase were immediately pooled and diluted with an equal volume of glycerol. TEVp-His6 was dialyzed into 20 mM 2-(N-morpholino)ethanesulfonic acid, 300 mM NaCl, 5% glycerol, 2 mM DTT, pH 6.0, quick frozen, and stored at −80 °C. TEVp-His6 was prepared at concentrations <2.5 mg/ml to avoid solubility problems.
Preparation of Pg species and fluorescent analogs of Pg
[Glu]Pg, [Lys]Pg , and Pm were prepared as previously described [5; 7; 21; 27; 31; 32]. Fluorescein-labeled [Glu]Pg and [Lys]Pg were prepared using SKΔ(R253-L260)ΔK414-His6. The thioester tripeptide chloromethyl ketone Nα-[(acetylthio)acetyl]-D-Phe-Phe-Arg-CH2Cl (ATA-FFR-CH2Cl) prepared as described [20] was used to inactivate SKΔ(R253-L260)ΔK414-His6·Pg* complex by adding 400 μM inhibitor to 25 μM of Pg in 1 M Hepes, 0.3 M NaCl, 1 mM EDTA, 1 mg/ml PEG-8000, pH 7.0 buffer. The reaction was initiated by adding 50 μM SKΔ(R253-L260)ΔK414-His6 and incubated at 25 °C for 2.5 h. The loss of activity was monitored from the initial rate of hydrolysis of D-Val-Leu-Lys-pNA (VLK-pNA) at 405 nm until it was <0.1% active. Excess inhibitor was removed by chromatography on Sephadex G-25 (fine) (1.5 cm × 30 cm) at 4 °C. All subsequent steps were done under reduced light. For labeling, 150 μM of 5-(iodoacetamido)fluorescein (5-IAF) was added to 6–15 μM SKΔ(R253-L260)ΔK414-His6·ATA-FFR-Pg* complex in the above pH 7.0 buffer and the reaction was initiated by addition of 1 M NH2OH to a final concentration of 0.1 M. After incubation at 25 °C for 1 h, 50 μM D-Phe-Phe-Arg-CH2Cl (FFR-CH2Cl) was added. The reaction mixture was then chromatographed on Sephadex G-25 (fine) (1.5 cm × 40 cm) at 22 °C to remove excess probe. The SKΔ(R253-L260)ΔK414-His6·labeled Pg* complex was dialyzed overnight in 0.1 M Hepes, 0.15 M NaCl, 10 mM 6-AHA, pH 7.4 buffer and then chromatographed on Ni2+-iminodiacetic acid-Sepharose to separate SKΔ(R253-L260)ΔK414-His6 from labeled Pg. Elution with a linear gradient of 3 M NaSCN over 6 column-volumes yielded labeled Pg with traces of labeled SK·Pg and SK·Pm complexes, and labeled Pm. Most of SKΔ(R253-L260)ΔK414-His6 remained bound to the Ni2+-Sepharose column and was eluted with 500 mM imidazole in 50 mM Hepes, 0.3 M NaCl, pH 7.4. An additional affinity chromatography step using native SK immobilized on AffiGel-10 (Bio-Rad) (1 cm × 16 cm; 5 mg coupled/ml of gel) separated labeled Pg from the SK·Pg/Pm complexes and labeled Pm. The complexes were eluted with 0.1 M Hepes, 0.125 M NaCl, 1 mM EDTA, 10 mM 6-AHA, pH 7.4 buffer. Elution with 20–25 column-volumes of a linear gradient up to 3 M NaSCN in the same buffer yielded labeled Pg in the first peak followed by labeled Pm. Labeled Pg was concentrated with a YM-30 ultrafiltration membrane and dialyzed against >5000 volumes of 50 mM Hepes, 125 mM NaCl, 1 mM EDTA, 1 mg/ml PEG 8000, pH 7.4. Quantitation of probe incorporation spectrophotometrically as described previously [20; 27; 30; 33] gave 0.86 ± 0.4 (mean and range) mol of probe/mol of protein for three preparations of [5F]FFR-[Lys]Pg, 0.94 ± 0.06 mol/mol (mean and range) for two preparations of [5F]FFR-[Glu]Pg, and similar values for other thiol-reactive probes. ATA-FFR-[Lys]Pg was labeled with Alexa Fluor 680 C2-maleimide (Invitrogen/Molecular Probes) ([AF680]FFR-[Lys]Pg) following the same procedure. The concentration of labeled Pg determined spectrophotometrically was confirmed by bicinchoninic acid protein micro-assay (Pierce).
Fluorescence equilibrium binding
Fluorescence was measured with an SLM 8100 spectrofluorometer in the ratio mode, using PEG 20,000-coated acrylic cuvettes. Experiments were performed in Buffer A containing 1 mM EDTA and 1 μM FFR-CH2Cl ± 10 mM 6-AHA as previously described [5; 7; 18; 27]. Fluorescence changes expressed as (Fobs-Fo)/Fo=ΔF/Fo, measured as a function of total SK species concentration, were fit by the quadratic binding equation, with the maximum fluorescence change ((Fmax-Fo)/Fo=ΔFmax/Fo), dissociation constant (Kd) the fitted parameters, and with the stoichiometric factor fixed at 1.
Plasminogen activation kinetics
Continuous assays of coupled conformational and proteolytic activation of [Lys]Pg by native SK, wtSK, wtSK-His6, and SKΔ(R253-L260)ΔK414-His6 were determined as described previously [5; 7; 27; 28]. Parabolic progress curves of hydrolysis of 200 μM VLK-pNA were obtained in the presence of 10 nM [Lys]Pg and increasing concentration of SK species. The first-order term in the second-order polynomial (Eq 1) fits represents the initial rate (v1) of VLK-pNA hydrolysis by the conformationally activated SK·Pg* complex, and the rate of activity increase (v2) obtained from the second-order term, reflects the initial rate of Pm generation.
| (Eq 1) |
The kinetics were analyzed for the simplified mechanism in Scheme 1, which assumes a negligible contribution from Pm generated by the SK·Pm complex [5; 7], where KA represents the dissociation constant for the SK·Pg* complex, and kPg the bimolecular rate constant (kcat/KS) for proteolytic generation of Pm by SK·Pg*. KA’ is the dissociation constant for the SK·Pm complex. Under the conditions of the experiments, where the concentration of free [Lys]Pg ([Pg]free) was much less than KS for [Lys]Pg binding to form the ternary enzyme-substrate SK·Pg*·Pg complex (270 nM), generation of Pm can be represented as a bimolecular reaction [5]. Under initial velocity conditions, the product is assumed to be SK·Pm because this complex is formed with a dissociation constant, KA’ of 12 pM, much tighter than [SK]o and [Pg]free [14]. When [SK]o>[Pg]o all of the product is SK·Pm, whereas when [SK]o<[Pg]o all of the SK becomes bound to Pm as the reaction progresses. v1 is given by Equation 2 where kcat/Km is the bimolecular rate constant for VLK-pNA hydrolysis by the SK·Pg* complex. For analysis of v1 as a function of SK species concentration, KA and kcat/Km were fitted parameters. v2 is given by Equation 3, where KI is the empirically determined competitive inhibition constant (330 ± 80 μM) of the bimolecular generation of Pm by the chromogenic substrate [5; 7]. [Pg]free is given by the quadratic Equation 4. For analysis of v2, KI was fixed at its previously determined value [5]. kPg and KA were the fitted parameters, where kPg is the bimolecular rate constant (kcat/KS in units of nM−1s−1) for Pm formation.
Scheme 1.
| (Eq 2) |
| (Eq 3) |
| (Eq 4) |
Nonlinear least-squares fitting was performed with SCIENTIST (MicroMath). Error estimates represent the 95% confidence interval.
Flow cytometry studies
Cells were isolated by Ficoll gradient and stained essentially as described previously [34; 35; 36]. Blood was collected by cardiac puncture. Splenocytes were isolated from whole spleens, which were filtered through 40 μm nylon mesh (BD Bioscience) to yield cellular suspensions. Cells were washed by repetitive centrifugation at 1500 rpm for 10 min at 4 °C with Dulbecco’s phosphate buffered saline (1 mM KH2PO4, 5.6 mM Na2HPO4, and 154 mM NaCl) supplemented with 0.5% bovine serum albumin and 1% fetal calf serum. Cells were incubated with an antibody cocktail against key immune cell surface markers, namely T cells (CD90-PE), B cells (B220-PE), NK cells (CD49b-PE and NK1.1-PE), granulocytes (Ly-6G-PE), residual red blood cells (Ter119-PE), and myeloid cells (CD11b-APC) (all from BD Biosciences). Cells were also simultaneously incubated with [AF680]FFR-[Lys]Pg to label Pg receptors on the immune cell surfaces. Competitive binding experiments with 6-AHA or native [Lys]Pg were performed in parallel tubes with competitors present in the cellular suspension just prior to addition of [AF680]FFR-[Lys]Pg. All reactions were incubated at 4 °C for 30 min prior to termination by 10-fold dilution with Dulbecco’s phosphate buffered saline supplemented with 0.5% bovine serum albumin and 1% fetal calf serum and centrifugation to recover cells. The cells were washed 2 additional times to remove excess antibodies, competitors, and [AF680]FFR-[Lys]Pg. Flow Cytometry data was collected on a LSRII Cytometer (BD Bioscience) after appropriate fluorescence compensations. Neutrophils were defined as CD11bhi(B220/CD90/CD49b/NK1.1./Ly6G/Ter119)hi and monocyte/macrophages as CD11bhi (B220/CD90/CD49b/NK1.1./Ly6G/Ter119)lo [34; 35; 36].
Results and discussion
A new approach to preparing active site-labeled Pg analogs
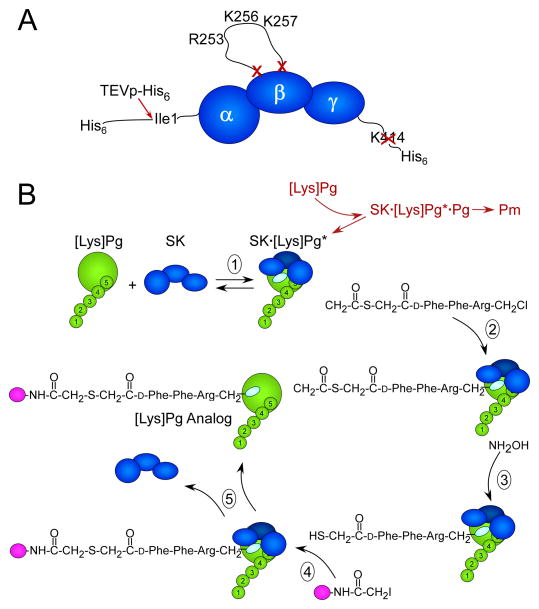
The identification of structural features of SK responsible for the Pg/Pm kringle LBS-mediated enhancement of SK·Pg* and SK·Pm catalytic complexes [27], and the LBS-dependent enhancement of Pg binding as a substrate of these complexes [28] prompted redesigning the method for preparation of active site-labeled Pg analogs. Initial attempts to use solely the His6-TEV-wtSK-His6 construct were successful, but limited by plasmin cleavage of SK to generate SK′, an NH2-terminal truncation of the first 59 residues, and a COOH-terminal cleavage that liberated the important His6-tag. The goal was to inhibit these coupled interactions, minimizing plasmin generation in the labeling reaction, while maintaining SK Ile1 required for conformational activation accompanying formation of SK·Pg*. To this end, a portion of the SK 250-loop containing the Arg253, Lys256, and Lys257 residues responsible for the LBS-dependent Pg substrate recognition was deleted [28] (Fig. 1A). The need to maintain SK Ile1 necessitated a TEVp cleavage site following the NH2-terminal His6-tag. A COOH-terminal His6-tag was added to replace the His6-tag lost after TEVp cleavage and to facilitate the removal of the SK bacterial activator from the labeled zymogen in the presence of NaSCN. Because the COOH-terminal Lys414 could be a Pm cleavage site, this residue was deleted, yielding the final construct, His6-(TEVp cleavage site)-SKΔ (R253-L260)ΔK414-His6 (Fig. 1A).
Figure 1. Illustrations of the SK construct used for active site-labeling and the labeling reaction scheme.
A. Modifications of SK to generate SKΔ(R253-L260)ΔK414-His6. SK represented (dark blue) by the α-, β-, and γ-domains from the NH2- to COOH-terminus. The NH2-terminal His6-tag is followed by the TEVp substrate recognition sequence (not shown) immediately adjacent to the essential Ile1- residue of the α-domain that is cleaved by TEVp-His6 (red arrow), removing the His6-segment. Residues Arg253-Leu260 in the SK β-domain containing the kringle 5-binding R253, K256, and K257 residues were deleted (red X, X). The COOH-terminal K414 was also deleted (red X). B. Reactions of the active site-labeling scheme: (1) SK (dark blue) binds to the [Lys]Pg (green) catalytic domain, which is attached to kringles 1–5 (numbered) forming the SK· [Lys]Pg* conformationally activated catalytic site (light blue oval). In coupled reactions (red) that are greatly inhibited by the use of the construct in A, [Lys]Pg binds as a substrate of SK· [Lys]Pg*, forming SK· [Lys]Pg*· [Lys]Pg, which is proteolytically activated to Pm. (2) Nα-[(acetylthio)acetyl]-D-Phe-Phe-Arg-CH2Cl reacts irreversibly with the active site in SK· [Lys]Pg*, trapping and inactivating the complex. (3) Reaction of the inhibitor incorporated into the SK· [Lys]Pg* active site with NH2OH generates a free thiol that in the presence of a fluorescence probe-iodoacetamide (magenta ball) (4) is selectively, covalently modified. (5) The probe-inhibitor-labeled SK· [Lys]Pg* complex is dissociated in NaSCN and the active site-labeled fluorescent [Lys]Pg analog is purified.
Separation of SKΔ (R253-L260)ΔK414-His6, His6-(TEVp cleavage site)-SKΔ (R253-L260)ΔK414-His6, and TEVp-His6 by Ni2+-iminodiacetic acid-Sepharose chromatography
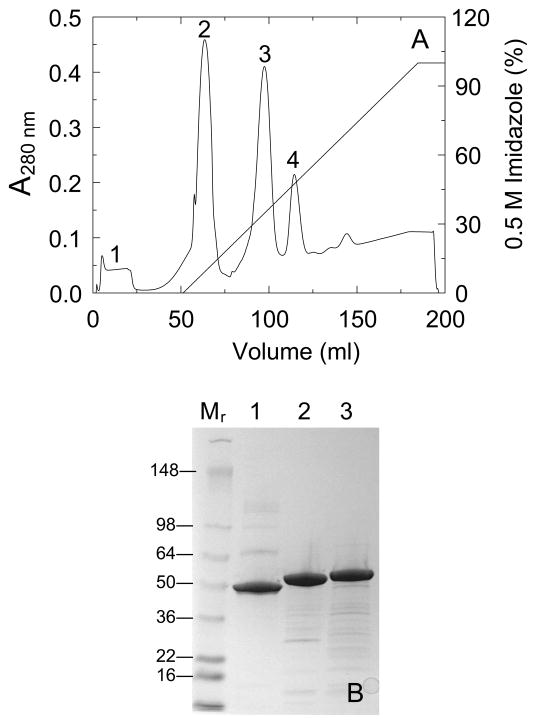
Fig. 2A shows a representative chromatogram of TEVp cleavage products generated from His6-(TEVp cleavage site)-SKΔ (R253-L260) ΔK414-His6 on Ni2+-iminodiacetic acid-Sepharose. Minor contaminants were eluted with the equilibration buffer containing 50 mM imidazole (peak 1). A shallow, linear gradient of imidazole up to 500 mM separated the TEVp-cleaved SKΔ (R253-L260) ΔK414-His6 (peak 2) from residual, uncleaved, double His6-tagged, His6-(TEVp cleavage site)-SKΔ (R253-L260) ΔK414-His6 (peak 3), and TEVp-His6 (peak 4). The His6-tagged SK constructs migrated at slightly higher apparent molecular mass on SDS-PAGE compared to native SK (Fig. 2B). The presence of SK Ile1 at the P1′ position3 of the TEVp substrate recognition sequence reduced the efficiency of TEVp cleavage to ~50% (see Materials and methods).
Figure 2. Purification of SKΔ (R253-L260) ΔK414-His6 by Ni2+- iminodiacetic acid-Sepharose chromatography.
A. Elution profile of the products of a reaction of His6-(TEVp cleavage site)-SKΔ (R253-L260) ΔK414-His6 with TEVp-His6 at a 10:1 substrate:protease molar ratio as described in Materials and methods, monitored by the A280 nm-absorbance eluting from a 5 ml Ni2+- iminodiacetic acid-Sepharose FPLC column equilibrated at 22 °C with 50 mM Hepes, 400 mM NaCl, 50 mM imidazole, pH 7.4, followed by a linear gradient of 0.05–0.5 M imidazole in the equilibration buffer. Minor contaminants eluting in the equilibration buffer (peak 1), the TEVp-cleaved protein SKΔ (R253-L260) ΔK414-His6 (peak 2), residual uncleaved His6-(TEVp cleavage site)-SKΔ (R253-L260) ΔK414-His6 (peak 3), and TEVp-His6 (peak 4). B. SDS-PAGE (4–15% gradient gel) of reduced samples (7 μg) of native SK (lane 1), SKΔ (R253-L260) ΔK414-His6 (lane 2), and His6-(TEVp cleavage site)-SKΔ (R253-L260) ΔK414-His6 (lane 3). Molecular mass markers in kDa (lane Mr).
Preparation of active site-labeled [Lys]Pg
Active site-labeled [Lys]Pg is difficult to purify because [Lys]Pg binds more tightly to SK than [Glu]Pg and is activated by SK faster to Pm than [Glu]Pg, although the affinity of SK·Pg* and Pg substrate binding are reduced in the new construct (see below). To reduce further the rate of Pm formation, the labeling reactions are typically carried out with a two-fold molar excess of SK. Although counterintuitive, this does not affect conformational activation, but inhibits the rate of Pm formation because Pg acts both as the catalyst and substrate. High concentrations of SK inhibit proteolytic activation by depletion of free Pg available to act as the substrate (see below and Scheme 1 in Materials and methods).
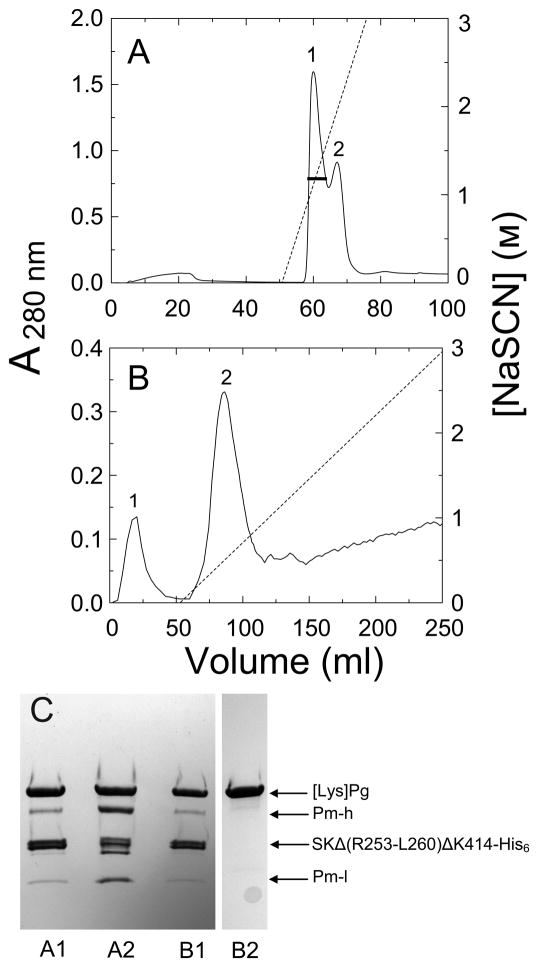
In the labeling scheme (Fig. 1B), [Lys]Pg in the presence of a large excess of ATA-FFR-CH2Cl was mixed with a 2-fold molar excess of SKΔ (R253-L260) ΔK414-His6 until the chromogenic substrate activity measured with VLK-pNA was <0.1%. Following removal of excess inhibitor by Sephadex G25 chromatography, the thiol was generated with NH2OH in the presence of excess 5-IAF, and excess probe removed by Sephadex G25 chromatography and dialysis. The labeled proteins were applied onto Ni2+-imminodiacetic acid-Sepharose eluted with a short gradient of NaSCN to dissociate labeled Pg, while the His-tagged SK mutant remained bound. This typically yields two poorly resolved peaks as shown by the elution profile in Fig. 3A and by SDS-PAGE (Fig. 3C). The first peak contains mostly labeled Pg, whereas the second contains more labeled Pm, which binds more tightly to the SK construct (Fig. 3C). The pooled fractions were dialyzed and loaded onto an SK-AffiGel-10 column, washed with the equilibration buffer, followed by a shallow gradient of NaSCN (Fig. 3B). As shown by SDS-PAGE (Fig. 3C), Pg and Pm bound to the SK construct passed through the column, and the purified [5F]FFR-[Lys]Pg was eluted at low NaSCN concentration, whereas any free labeled Pm eluted at higher NaSCN concentration (not visible in this preparation).
Figure 3. Purification of labeled [Lys]Pg from labeling reaction mixtures by Ni2+-iminodiacetic acid-Sepharose and SK-AffiGel-10 affinity chromatography.
A. Elution profile, measured by A280 nm-absorbance, of [Lys]Pg products from a reaction mixture of 25 μM [Lys]Pg with 50 μM SKΔ (R253-L260) ΔK414-His6 inactivated with 410 μM ATA-FFR-CH2Cl, and subsequently labeled with 155 μM 5-IAF on Ni2+- iminodiacetic acid-Sepharose (5 ml). Proteins were eluted with a steep gradient of NaSCN over 6 column-volumes. The black bar represents the fractions of peak 1 pooled. B. Elution profile of the pooled, concentrated, and dialyzed fractions in A on SK-AffiGel-10 (1 cm 16 cm; 5 mg of SK/ml of gel) as described in Materials and methods. The SK mutant·Pg/Pm complexes elute in the equilibration buffer (peak 1), labeled [Lys]Pg (peak 2) is eluted with a shallow gradient of 3 M NaSCN in the equilibration buffer over 20 column-volumes, and labeled Pm elutes at higher NaSCN (not detectable in this chromatogram). C. SDS-PAGE (4–15% gradient gels) of reduced samples (8–10 μg) from panel A, peak 1 (A1) and peak 2 (A2), and from panel B, peak 1 (B1) and peak 2 (B2). Sample B2 was from the same preparation run on a separate, identical gel. Bands representing labeled [Lys]Pg ([Lys]Pg), SKΔ (R253-L260) ΔK414-His6 (SKΔ (R253-L260) ΔK414-His6), the Pm heavy chain (Pm-h) and labeled light chain (Pm-l) are identified by the arrows.
The yield of [5F]FFR-[Lys]Pg from this representative preparation was 1.1 mg, 13% of the starting 8.4 mg of [Lys]Pg, representing a 2.6-fold improvement in the yield compared that of the previous method of ~5%. The average yield from 4 preparations of labeled [Lys]Pg is 13%. The same procedure was used for preparation of [5F]FFR-[Glu]Pg, with an average yield of 20% from two preparations. The time required for the improved method is about half of that for the previous procedure and only requires preparation of one affinity column instead of two [7; 16]
Specificity of active site-labeling of [Glu]Pg
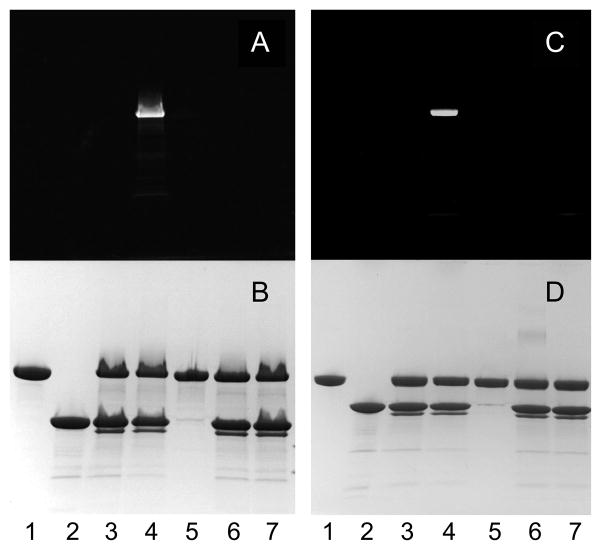
To confirm the active site specificity of labeling by the new method, incorporation 5-IAF in a small-scale set of [Glu]Pg labeling reactions using SKΔ (R253-L260)ΔK414-His6 and ATA-FFR-CH2Cl was assessed by SDS-PAGE (Fig. 4; see Fig 1B). [Glu]Pg incubated with ATA-FFR-CH2Cl and SKΔ(R253-L260)ΔK414-His6 followed by removal of excess inhibitor and reaction with 5-IAF in the presence of NH2OH showed covalent incorporation of fluorescein into the Pg zymogen (Fig. 4A and 4B, lanes 4). Reactions in the absence of the SK mutant showed no labeling (lanes 5). Omitting NH2OH to generate the thiol, or blocking the active site with FFR-CH2Cl before subjection to ATA-FFR-CH2Cl in the presence of SKΔ (R253-L260) ΔK414-His6, and subsequently, 5-IAF in the presence of NH2OH both showed no fluorescein incorporation (lanes 6 and 7, respectively; see Fig. 1B). In all reactions containing either ATA-FFR-CH2Cl or FFR-CH2Cl and the SK construct, a band migrating slightly lower than the SK construct on SDS-PAGE was generated, which likely represents SK′. SK′ is produced by Pm cleavage at Lys59 in the SK α-domain [37; 38] The SK-(1–59) peptide remains bound to the remainder of the SK α-domain, which retains its folded structure under non-denaturing conditions [37; 39; 40]. SK′ has approximately the same affinity as native SK for Pm [14].
Figure 4. Specificity of active site-labeling of conformationally activated SKΔ (R253-L260) Δ K414-His6· [Glu]Pg* complex with ATA-FFR-CH2Cl and 5-IAF assessed by SDS-PAGE.
The fluorescence (A and C) and protein-stained bands (B and D) are shown for reduced (A and B) and non-reduced (C and D) samples (~11 μg). Reactions were performed with 15 μM [Glu]Pg, 30 μM SKΔ (R253-L260) ΔK414-His6, and 355 μM ATA-FFR-CH2Cl for 2 h at 25 °C, followed by removal of excess inhibitor by Sephadex G25 centrifugal chromatography through 2 ml spin-columns (Bio-Rad). Free thiol was generated by addition of 1 M NH2OH to 0.1 M and incubation with 170 μM 5-IAF for 2 h at 25 °C . Free dye was removed by Sephadex G25 (2 ml) centrifugal chromatography. [Glu]Pg (lane1), SKΔ (R253-L260) Δ K414-His6 (lane 2), the SKΔ (R253-L260) ΔK414-His6· [Glu]Pg* complex after inactivation with ATA-FFR-CH2Cl (lane 3), the complete set of labeling reactions (lane 4), control reactions where SKΔ (R253-L260) ΔK414-His6 was omitted (lane 5), NH2OH was omitted (lane 6), or the catalytic site of SKΔ (R253-L260) ΔK414-His6· [Glu]Pg* complex was blocked with 200 μM FFR-CH2Cl before incubation with ATA-FFR-CH2Cl, and 5-IAF in the presence of NH2OH, as described above (lane 7).
Equilibrium binding of native SK, wtSK-His6, and SKΔ (R253-L260)ΔK414-His6 to fluorescein-labeled [Lys]Pg
The affinity of native SK, wtSK-His6, and SKΔ (R253-L260) ΔK414-His6 for binding to [5F]FFR-[Lys]Pg was determined in titrations of the fluorescence quenching accompanying the interaction [7; 16]. As shown previously, this measures the affinity for labeled Pg binding in the catalytic mode to SK [7]. A consistent property of the active site-labeled Pg analogs (not labeled Pm) is that the affinity is ~5-fold weaker than of unlabeled Pg, reflecting changes in the conformation of the Pg catalytic domain due to occupation of the active site by the probe-tripeptide [7; 15]. The analogs retain the dependence of affinity on LBS interactions.
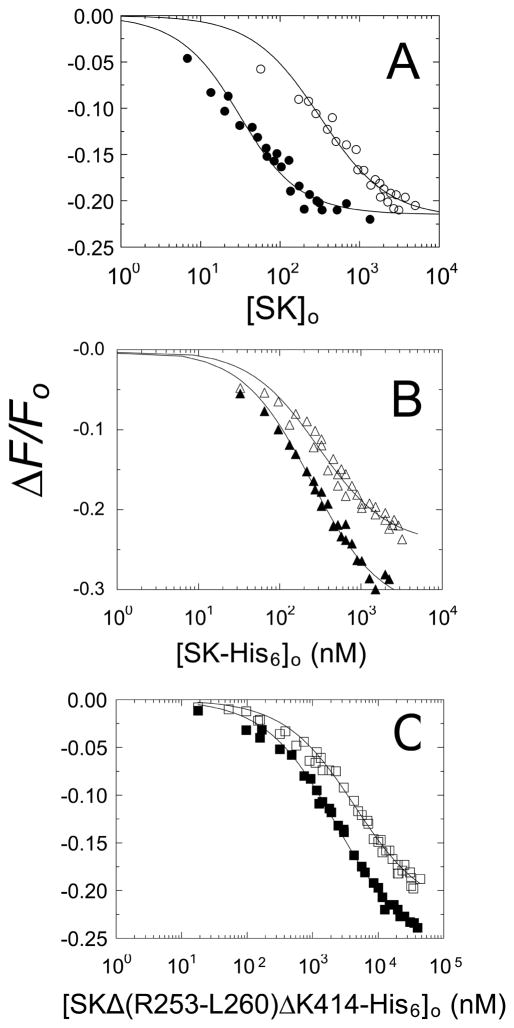
Titrations of [5F]FFR-[Lys]Pg with native SK and wtSK-His6 were performed in the absence and presence of 10 mM 6-AHA (Fig. 5A and 5B). In the absence and presence of 6-AHA, native SK bound labeled [Lys]Pg with Kd 23 ± 6 nM and 310 ± 70 nM, respectively (Fig. 5A; Table 1). Consistent with our previous studies of an SK mutant lacking the COOH-terminal lysine [27], wtSK-His6 bound to labeled [Lys]Pg with weaker affinity than native SK, Kd 240 ± 30 nM and 290 ± 50 nM in the absence and presence of 6-AHA, respectively (Fig. 5B). Blockage of the COOH-terminal lysine residue in wtSK-His6 decreased the weakening effect of 6-AHA on affinity from 13.5 fold to 1.2-fold (Table 1). This demonstrated that the SK mutant with an obstructed COOH-terminal lysine binds labeled [Lys]Pg without the normal increase in affinity due to LBS interaction with a Pg kringle. Similarly, SKΔ (R253-L260) ΔK414-His6 exhibited a 20-fold lower affinity than native SK in the absence of 6-AHA and 2.6-fold lower in the presence of 10 mM 6-AHA (Fig. 5C; Table 1).
Figure 5. Effects of an SK COOH-terminal His6-tag and deletion of Arg253-Leu260 on the affinity of SK for [5F]FFR-[Lys]Pg.
A. Fluorescence titrations (ΔF/Fo) of 15 nM [5F]FFR-[Lys]Pg as a function of total SK concentration ([SK]o) for native SK in the absence (●) and presence (○) of 10 mM 6-AHA. B. Titrations as in A with SK-His6 in the absence (▲) and presence (△) of 10 mM 6-AHA. C. Titrations as in A with SKΔ (R253-L260) ΔK414-His6 in the absence (■) and presence (□) of 10 mM 6-AHA. Solid lines represent the non-linear least-squares fits to the data by the quadratic binding equation with the parameters in Table 1. Titrations were performed and analyzed as described in Materials and methods.
Table 1.
Equilibrium binding parameters for SK constructs binding to 5-fluorescein-labeled [Lys]Pga
| SK construct | No 6-AHA
|
10 mM 6-AHA
|
||
|---|---|---|---|---|
| Kd (nM) | ΔFmax/Fo (%) | Kd (nM) | ΔFmax/Fo (%) | |
| Native SK | 23 ± 6b | −22 ± 1 | 310 ± 60 | −22 ± 1 |
| SK-His6 | 240 ± 30 | −33 ± 1 | 290 ± 50 | −24 ± 1 |
| SKΔ (R253-L260) ΔK414-His6 | 470 ± 40 | −26 ± 1 | 820 ± 90 | −22 ± 1 |
Dissociation constants (Kd) and maximum fluorescence changes (ΔFmax/Fo) are listed for the titrations shown in Fig. 5. [5F]FFR-[Lys]Pg (15 nM) was titrated with the indicated SK constructs in the absence (No 6-AHA) and presence of 6-AHA (10 mM 6-AHA). Binding experiments were performed and analyzed as described in Materials and methods.
Experimental error represents the 95% confidence interval.
Kinetics of conformational and proteolytic [Lys]Pg activation by SK mutants
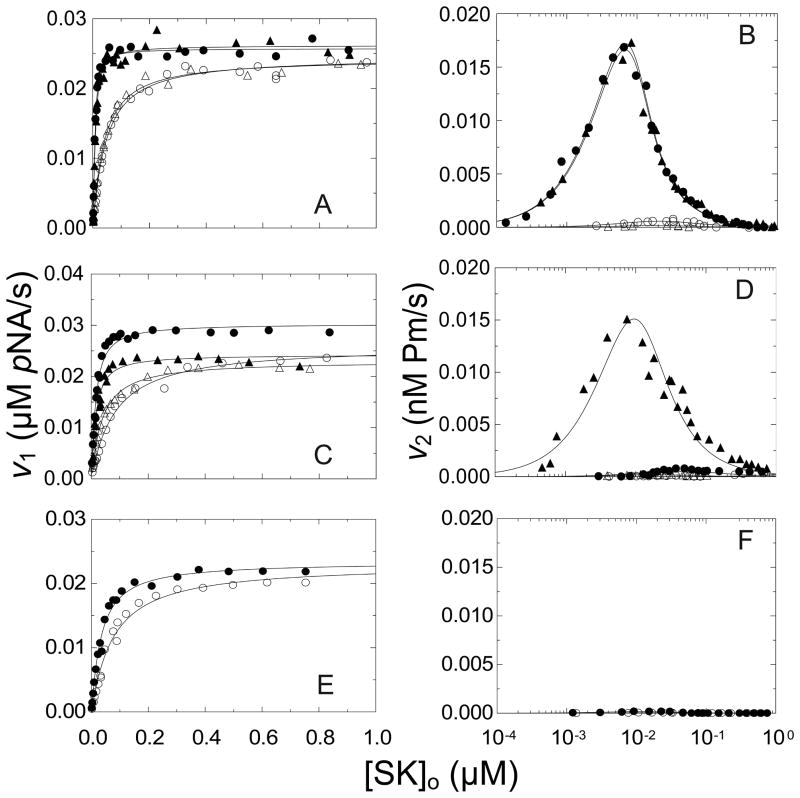
SK mutants were characterized further in kinetic studies of [Lys]Pg activation. The results were analyzed using a kinetic model shown in Scheme 1 (Materials and methods). The kinetics of [Lys]Pg activation were monitored by hydrolysis of VLK-pNA and the parabolic progress curves were resolved into an initial rate of substrate hydrolysis (v1) representing the activity of the rapidly formed, conformationally activated SK· [Lys]Pg* complex, and the rate of acceleration (v2) representing binding of [Lys]Pg as a substrate of the SK· [Lys]Pg* complex and its proteolytic conversion to Pm [5; 7]. The SK concentration dependence of v1 is hyperbolic, yielding the dissociation constant for SK·Pg* complex (KA(v1)) and the maximum, representing the bimolecular rate constant for chromogenic substrate hydrolysis by this complex (kcat/Km(v1)). v2 shows behavior unique to the SK mechanism, due to Pg binding to SK in both catalytic mode and substrate mode (see Scheme 1 in Materials and methods). For [Lys]Pg, which binds SK tightly in the catalytic mode, v2 increases sharply to a maximum at [SK]o ≈ 0.5[Pg]o and decreases at higher SK concentration, approaching zero when [SK]o ≫[Pg]o due to depletion of free [Lys]Pg to act as substrate [5]. Because the reactions are coupled, analysis of the SK dependences of v1 and v2 both provide an estimate of the SK· [Lys]Pg* dissociation constant (KA(v1) and KA(v2)). KA(v1) is generally more reliable because generation of free Pm that binds more tightly to SK skews the estimate of KA(v2)) toward lower values [5]. Analysis of the SK concentration dependence of v2 also yields the bimolecular rate constant for [Lys]Pg conversion to Pm (kPg).
Parameters derived from v1 and v2 in the absence, and v1 in the presence of 10 mM 6-AHA are summarized in Table 2. It should be noted that because v2 is vastly inhibited by 10 mM 6-AHA, no parameters for Pm generation were obtained under these conditions. Native and wtSK showed indistinguishable KA(v1) of 2 ± 1 and 4 ± 1 nM in the absence of 6-AHA, respectively, and 20- or 8-fold weaker values, respectively, in the presence of 6-AHA (Fig. 6A and 6B). The bimolecular rate constant for VLK-pNA hydrolysis was similar for native and wtSK, as was kPg (Table 2).
Table 2.
Kinetic parameters for conformational and proteolytic activation of [Lys]Pg by SK constructsa
| SK construct | No 6-AHA
|
10 mM 6-AHA
|
||||
|---|---|---|---|---|---|---|
| KA (v1) (nM) | kcat/Km (v1) (nM−1s−1) | KA (v2) (nM) | kPg (v2) (nM−1s−1) | KA (v1) (nM) | kcat/Km (v1) (nM−1s−1) | |
| Native SK | 2 ± 1b | 13.2 ± 0.4 | 1.6 ± 0.2 | 1020 ± 40 | 40 ± 5 | 12.6 ± 0.3 |
| wild-type SK | 4 ± 1 | 13.4 ± 0.5 | 1.6 ± 0.2 | 1010 ± 40 | 33 ± 4 | 12.5 ± 0.3 |
| SKΔ(R253-L260) | 10 ± 2 | 15.5 ± 0.5 | NAc | NA | 78 ± 13 | 13.3 ± 0.7 |
| SK-His6 | 10 ± 4 | 12.5 ± 0.7 | 4 ± 1 | 900 ± 40 | 32 ± 3 | 11.8 ± 0.3 |
| SKΔ(R253-L260)ΔK414-His6 | 28 ± 5 | 12.0 ± 0.5 | NA | NA | 70 ± 7 | 11.8 ± 0.7 |
The apparent dissociation constant for conformational activation (KA (v1)) of 10 nM [Lys]Pg by the indicated SK constructs, and the bimolecular rate constant for VLK-pNA hydrolysis by the SK·[Lys]Pg* complex (kcat/Km (v1)) in the absence (No 6-AHA) and presence of 6-AHA (10 mM 6-AHA). Also listed are KA obtained from v2 (KA (v2)) and the bimolecular rate constant for Pm generation from [Lys]Pg (kPg). Parameters were determined by analysis of the data shown in Fig. 6 as described in Materials and methods.
Experimental error in the parameters represents the 95% confidence interval.
NA represents not analyzable
Figure 6. Steady-state kinetics of conformational and proteolytic [Lys]Pg activation by SK constructs.
A and B. v1 and v2, respectively, as a function of total SK concentration ([SK]o) for native SK (●) and wtSK (▲). C and D, v1 and v2, respectively, for SKΔ(R253-L260) (●) and wtSK-His6 (▲). E. and F., v1 and v2, respectively, for SKΔ(R253-L260))ΔK414-His6 (●). Experiments were performed in the absence solid symbols (●,▲) or presence open symbols (○,△) of 10 mM 6-AHA. The solid lines represent the fits of Eq 2 and 4 for v1, and Eq 3 and 4 for v2 in Materials and methods with the parameters listed in Table 2. Kinetics of 10 nM [Lys]Pg activation by SK constructs were performed and analyzed as described in Materials and methods.
SKΔ(R253-L260) and SK-His6 both showed 2.5-fold lower affinity (KA(v1) 10 nM) for conformational activation compared to wtSK, which was reduced 8- and 3-fold, respectively by 10 mM 6-AHA (Fig. 6C; Table 2). SK-His6 exhibited kPg indistinguishable from wtSK, whereas SKΔ(R253-L260) showed slow rates of Pm formation that could not be analyzed (Fig. 6D). The near-absence of Pm generation with SKΔ(R253-L260) was consistent with previous results with the SKΔ (R253-L260)ΔK414 mutant, which lacks the LBS-dependence of Pg substrate recognition [28]. The SKΔ (R253-L260) ΔK414-His6 mutant had KA(v1) of 28 ± 5 nM, 7-fold weaker than wtSK, which was increased only 2.5-fold by 10 mM 6-AHA (Fig. 6E and 6F; Table 2). As expected, only slow rates of Pm formation were observed with this mutant. The results of the kinetic studies paralleled those of the equilibrium binding experiments in that blocking Lys414 in SK-His6 or deletion of Lys414 and blocking the COOH-terminus in SKΔ(R253-L260) ΔK414-His6 nearly eliminated the effects of 6-AHA on both SK-[Lys]Pg binding and formation of the conformationally activated SK· [Lys]Pg* complex. In the binding studies, the 13.5-fold weakening effect of 6-AHA on native SK affinity for [Lys]Pg was reduced for SK-His6 and SKΔ (R253-L260) ΔK414-His6 to 1.2- and 1.7-fold, respectively (Table 1). Similarly, the effect of 6-AHA on the kinetically determined KA(v1) for native SK reduced the 20-fold effect of 6-AHA on SK· [Lys]Pg* formation to 3- and 2.5-fold for the two constructs, respectively (Table 2) [27].
Application of the labeling method to quantify [Lys]Pg receptors on leukocytes using the near-IR probe Alexa Fluor 680
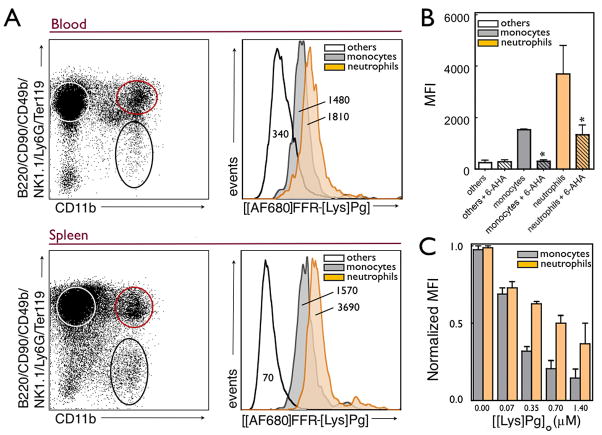
Because there are a number of Pg receptors, including α-enolase, annexin II [41; 42], p11 [43], Plg-RKT [44], and histone H2B [45] present on cell surfaces, we sought to develop a probe that would assess the overall level of Pg binding capacity of cells from a complex biologic mixture. [Lys]Pg was chosen as the receptor ligand, in part, because it has been shown that on endothelial cells and monocytes, [Glu]Pg is rapidly converted to [Lys]Pg [46; 47]. Many Pg receptors mediate binding via the LBS in kringle domains and it is well known that [Lys]Pg expresses enhanced kringle LBS interactions compared to [Glu]Pg [3; 16; 48; 49; 50], further favoring the choice of [Lys]Pg. Flow cytometry analysis of immune cells isolated from either murine blood or spleen showed [Alexa Fluor 680]FFR-[Lys]Pg ([AF680]FFR-[Lys]Pg) labeling of primarily CD11b+ cells, namely neutrophils and monocytes. The flow cytometry gating strategy exploited here, B220/CD90/CD49b/NK1.1/Ly6G/Ter119 versus CD11b+, has been documented and is a well accepted protocol [34; 35; 36]. Populations of cells were confirmed by cytospin morphologic analysis after H&E staining (data not shown). Results from labeled splenocytes are shown in Fig. 7A and indicate that [AF680]FFR-[Lys]Pg labels CD11b+ cells (monocytes/macrophages and neutrophils) preferentially over lymphocytes, indicated by an average of 38-fold higher mean fluorescence intensity (MFI). This was also observed when we stained cells obtained from blood, but the difference between cell populations was less pronounced (Fig. 7A). Neutrophils had slightly higher [AF680]FFR-[Lys]Pg positive values as demonstrated by the representative MFI histograms. These labeling results were similar in both cells harvested from blood and from the spleen. Binding of [AF680]FFR-[Lys]Pg to CD11b+ cells was blocked by the addition of 5 mM 6-AHA (Fig. 7B). To validate that the fluorescence signal demonstrated by populations of neutrophils and monocytes resulted from surface labeling of [AF680]FFR-[Lys]Pg, a competitive binding experiment was conducted between native [Lys]Pg and [AF680]FFR-[Lys]Pg (Fig. 7C). The MFI of the neutrophil and monocyte groups decreased in a concentration-dependent manner with increasing [Lys]Pg. The results demonstrate the ability of labeled [Lys]Pg to be used in combination with other immune markers to identify specific populations of cells and assess the expression of Pg receptors on the surface of these cells. The advantage of this strategy over antibody staining is two-fold: first, antibody staining for a single Pg receptor on the cell surface would grossly underestimate the total Pg binding potential; and second, the approach allows specific incorporation of near-IR probes or other molecular beacons. Such probes could potentially be used for whole animal imaging of Pg-positive migrating cells [45; 51; 52; 53].
Figure 7. Application of the active site-labeling procedure to flow cytometry analysis of Pg receptors on immune cells visualized with [AlexaFluor 680]FFR-[Lys]Pg.
A, Representative flow cytometry dot plots collected from the blood (top) and spleen (bottom) from C57BL/6 mice (n=3). Populations of cells studied are highlighted with others cells (lymphocytes and red blood cells) denoted by the white circle, neutrophils (CD11b+ cells, but positive for B220/CD90/CD49b/NK1.1/Ly6G/Ter119 antibody cocktail) by the orange circle and monocytes (CD11b+ and B220/CD90/CD49b/NK1.1/Ly6G/Ter119 negative) by the black circle. Total [AF680]FFR-[Lys]Pg uptake for these same immune cell groups is shown as histograms with the MFI shown for each. B. [AF680]FFR-[Lys]Pg MFI compared among the same groups isolated from the spleen after incubation with or without 5 mM 6-AHA at 4 °C for 30 min. C. Binding of [AF680]FFR-[Lys]Pg to CD11b+ cells in the presence of increasing concentrations of [Lys]Pg. Results demonstrate the specificity of [AF680]FFR-[Lys]Pg labeling for labeling of neutrophils and monocytes. Experiments were performed in triplicate as described in Materials and methods, and standard error of the mean is shown. * indicates p < 0.05.
Pg plays a critical role in intravascular fibrinolysis mainly through its activation by tissue-type Pg activator in association with annexin II on endothelial cells, monocytes, and macrophages, which are involved in cell migration, wound repair, and tissue remodeling [51; 53; 54; 55]. In addition, extravascular Pg, estimated to be 40% of total Pg [56], is activated mainly by urokinase-type Pg activator and its receptor, urokinase-type Pg activator receptor (uPAR), which has an established role in tumor invasion and metastasis of a number of cancers by generating Pm that breaks down extracellular matrix and facilitates metalloproteinase activation [56; 57; 58; 59; 60; 61]. This has led to sophisticated approaches for specific imaging of uPA activity (e.g. [62]). Fewer studies have addressed Pg binding to its numerous receptors [52; 61; 63]. Our studies of generic detection of [Lys]Pg-binding receptors provide a platform for new receptor-specific imaging of Pg/Pm on normal migrating cells and on cells in a variety of pathological settings. With respect to cancer imaging, the challenge is whether the differential imaging of normal monocytes/macrophages and these cells in cancer metastasis is large enough to be used diagnostically.
Conclusions
An SK mutant was rationally engineered that exhibits greatly reduced Pm generation, while maintaining sufficient affinity for Pg conformational activation to facilitate the production of active site-specific and stoichiometrically labeled fluorescent Pg analogs. Use of the SK mutant increased the yield for labeling of [Lys]Pg and [Glu]Pg from ~5% to 13% and ~20%, respectively, reduced the time of their preparation at least 2-fold, and maintained the high purity of previous methods. The versatility of the labeling scheme was advanced to include application of [Lys]Pg labeled with the near-infrared probe, Alexa Fluor 680, to identify and quantitate murine immune cell populations harboring Pg receptors by flow cytometry. The results suggest that new Pg analogs may be designed by the same strategy for imaging of normal and pathological cellular functions.
Acknowledgments
We thank Dr. Laura Mizoue of the Vanderbilt University Center for Structural Biology for the TEV proteinase construct, and Dr. Jonathan Creamer for his work optimizing the TEV proteinase purification. We thank Dr. Virna Cortez-Retamozo and Dr. Cedric Berger for help acquiring the flow cytometry data. We also thank the Molecular Structure Facility at the University of California at Davis for performing NH2-terminal sequencing and the Vanderbilt DNA Sequencing Facility for DNA sequencing.
Footnotes
This work was supported in part by National Institutes of Health Grant RO1 HL056181 from the National Heart, Lung, and Blood Institute to P. E. B., F32 HL094010 and K99 HL094533 to P.P., R01 HL096576-01 and an American Heart Association grant (0835623D) to M.N. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health. The funding organizations played no role in the decision to publish or the content.
Abbreviations used: SK, streptokinase; wtSK, recombinant wild-type SK; SKΔK414, SK lacking the COOH-terminal Lys414 residue; SKΔ(R253-L260), SK with residues Arg253 to Leu260 deleted; SKΔ (R253-L260) ΔK414, SK with residues Arg253 to Leu260 and Lys414 deleted; [Glu]Pg, intact native plasminogen; [Lys]Pg, native Pg lacking the NH2-terminal 77 residues; Pm, plasmin; μPm;, the Pm catalytic domain; FFR-CH2Cl, D-Phe-Phe-Arg-CH2Cl; ATA-FFR-CH2Cl, Nα-[(acetylthio)acetyl]-D-Phe-Phe-Arg-CH2Cl; 5-IAF, 5-(iodoacetamido)fluorescein; 6-AHA, 6-aminohexanoic acid; Pg*, non-proteolytically activated form of the plasminogen zymogen; LBS, lysine-binding sites; PEG, polyethylene glycol; pNA, para-nitroaniline: VLK-pNA, D-Val-Leu-Lys-pNA; [5F]FFR-[Lys]Pg or [5F]FFR-[Glu]Pg, [Lys]Pg or [Glu]Pg inactivated with ATA-FFR-CH2Cl and labeled with 5-IAF; [AF680]FFR-[Lys]Pg, [Lys]Pg inactivated with ATA-FFR-CH2Cl and labeled with Alexa Fluor 680 C2-maleimide; MFI, mean fluorescence intensity; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide-gel electrophoresis; TEVp, tobacco etch virus proteinase.
Schechter-Berger [64] notation referring to the residues of a substrate (from the NH2-terminal end) as …P4-P3-P2-P1-P1′-P2′… with the scissile bond at P1-P1′.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown PJ, Mulvey D, Potts JR, Tomley FM, Campbell ID. Solution structure of a PAN module from the apicomplexan parasite Eimeria tenella. J Struct Funct Genomics. 2003;4:227–34. doi: 10.1023/b:jsfg.0000016118.87333.03. [DOI] [PubMed] [Google Scholar]
- 2.Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461:63–7. doi: 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- 3.Castellino FJ, McCance SG. The kringle domains of human plasminogen. Ciba Found Symp. 1997;212:46–60. doi: 10.1002/9780470515457.ch4. [DOI] [PubMed] [Google Scholar]
- 4.Ponting CP, Marshall JM, Cederholm-Williams SA. Plasminogen: a structural review. Blood Coagul Fibrinolysis. 1992;3:605–14. [PubMed] [Google Scholar]
- 5.Boxrud PD, Bock PE. Coupling of conformational and proteolytic activation in the kinetic mechanism of plasminogen activation by streptokinase. J Biol Chem. 2004;279:36642–9. doi: 10.1074/jbc.M405265200. [DOI] [PubMed] [Google Scholar]
- 6.Chibber BA, Morris JP, Castellino FJ. Effects of human fibrinogen and its cleavage products on activation of human plasminogen by streptokinase. Biochemistry. 1985;24:3429–34. doi: 10.1021/bi00335a006. [DOI] [PubMed] [Google Scholar]
- 7.Boxrud PD, Verhamme IM, Bock PE. Resolution of conformational activation in the kinetic mechanism of plasminogen activation by streptokinase. J Biol Chem. 2004;279:36633–41. doi: 10.1074/jbc.M405264200. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Lin X, Loy JA, Tang J, Zhang XC. Crystal structure of the catalytic domain of human plasmin complexed with streptokinase. Science. 1998;281:1662–5. doi: 10.1126/science.281.5383.1662. [DOI] [PubMed] [Google Scholar]
- 9.Teuten AJ, Broadhurst RW, Smith RA, Dobson CM. Characterization of structural and folding properties of streptokinase by n.m.r. spectroscopy. Biochem J. 1993;290(Pt 2):313–9. doi: 10.1042/bj2900313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damaschun G, Damaschun H, Gast K, Gerlach D, Misselwitz R, Welfle H, Zirwer D. Streptokinase is a flexible multi-domain protein. Eur Biophys J. 1992;20:355–61. doi: 10.1007/BF00196594. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Reed GL, Hedstrom L. Deletion of Ile1 changes the mechanism of streptokinase: evidence for the molecular sexuality hypothesis. Biochemistry. 1999;38:5232–40. doi: 10.1021/bi981915h. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Reed GL, Hedstrom L. Zymogen activation in the streptokinase-plasminogen complex. Ile1 is required for the formation of a functional active site. Eur J Biochem. 2000;267:3994–4001. doi: 10.1046/j.1432-1327.2000.01434.x. [DOI] [PubMed] [Google Scholar]
- 13.Boxrud PD, Verhamme IM, Fay WP, Bock PE. Streptokinase triggers conformational activation of plasminogen through specific interactions of the amino-terminal sequence and stabilizes the active zymogen conformation. J Biol Chem. 2001;276:26084–9. doi: 10.1074/jbc.M101966200. [DOI] [PubMed] [Google Scholar]
- 14.Boxrud PD, Fay WP, Bock PE. Streptokinase binds to human plasmin with high affinity, perturbs the plasmin active site, and induces expression of a substrate recognition exosite for plasminogen. J Biol Chem. 2000;275:14579–89. doi: 10.1074/jbc.275.19.14579. [DOI] [PubMed] [Google Scholar]
- 15.Bock PE, Day DE, Verhamme IM, Bernardo MM, Olson ST, Shore JD. Analogs of human plasminogen that are labeled with fluorescence probes at the catalytic site of the zymogen. Preparation, characterization, and interaction with streptokinase. J Biol Chem. 1996;271:1072–80. doi: 10.1074/jbc.271.2.1072. [DOI] [PubMed] [Google Scholar]
- 16.Boxrud PD, Bock PE. Streptokinase binds preferentially to the extended conformation of plasminogen through lysine binding site and catalytic domain interactions. Biochemistry. 2000;39:13974–81. doi: 10.1021/bi000594i. [DOI] [PubMed] [Google Scholar]
- 17.Bock PE. Thioester peptide chloromethyl ketones: reagents for active site-selective labeling of serine proteinases with spectroscopic probes. Methods Enzymol. 1993;222:478–503. doi: 10.1016/0076-6879(93)22030-j. [DOI] [PubMed] [Google Scholar]
- 18.Bean RR, Verhamme IM, Bock PE. Role of the streptokinase alpha-domain in the interactions of streptokinase with plasminogen and plasmin. J Biol Chem. 2005;280:7504–10. doi: 10.1074/jbc.M411637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verhamme IM, Olson ST, Tollefsen DM, Bock PE. Binding of exosite ligands to human thrombin. Re-evaluation of allosteric linkage between thrombin exosites I and II. J Biol Chem. 2002;277:6788–98. doi: 10.1074/jbc.M110257200. [DOI] [PubMed] [Google Scholar]
- 20.Bock PE. Active-site-selective labeling of blood coagulation proteinases with fluorescence probes by the use of thioester peptide chloromethyl ketones. II. Properties of thrombin derivatives as reporters of prothrombin fragment 2 binding and specificity of the labeling approach for other proteinases. J Biol Chem. 1992;267:14974–81. [PubMed] [Google Scholar]
- 21.Dharmawardana KR, Bock PE. Demonstration of exosite I-dependent interactions of thrombin with human factor V and factor Va involving the factor Va heavy chain: analysis by affinity chromatography employing a novel method for active-site-selective immobilization of serine proteinases. Biochemistry. 1998;37:13143–52. doi: 10.1021/bi9812165. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003;425:535–9. doi: 10.1038/nature01962. [DOI] [PubMed] [Google Scholar]
- 23.Lin LF, Houng A, Reed GL. Epsilon amino caproic acid inhibits streptokinase-plasminogen activator complex formation and substrate binding through kringle-dependent mechanisms. Biochemistry. 2000;39:4740–5. doi: 10.1021/bi992028x. [DOI] [PubMed] [Google Scholar]
- 24.Conejero-Lara F, Parrado J, Azuaga AI, Dobson CM, Ponting CP. Analysis of the interactions between streptokinase domains and human plasminogen. Protein Sci. 1998;7:2190–9. doi: 10.1002/pro.5560071017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhar J, Pande AH, Sundram V, Nanda JS, Mande SC, Sahni G. Involvement of a nine-residue loop of streptokinase in the generation of macromolecular substrate specificity by the activator complex through interaction with substrate kringle domains. J Biol Chem. 2002;277:13257–67. doi: 10.1074/jbc.M108422200. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhary A, Vasudha S, Rajagopal K, Komath SS, Garg N, Yadav M, Mande SC, Sahni G. Function of the central domain of streptokinase in substrate plasminogen docking and processing revealed by site-directed mutagenesis. Protein Sci. 1999;8:2791–805. doi: 10.1110/ps.8.12.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panizzi P, Boxrud PD, Verhamme IM, Bock PE. Binding of the COOH-terminal lysine residue of streptokinase to plasmin(ogen) kringles enhances formation of the streptokinase.plasmin(ogen) catalytic complexes. J Biol Chem. 2006;281:26774–8. doi: 10.1074/jbc.C600171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tharp AC, Laha M, Panizzi P, Thompson MW, Fuentes-Prior P, Bock PE. Plasminogen substrate recognition by the streptokinase-plasminogen catalytic complex is facilitated by Arg253, Lys256, and Lys257 in the streptokinase beta-domain and kringle 5 of the substrate. J Biol Chem. 2009;284:19511–21. doi: 10.1074/jbc.M109.005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapust RB, Tozser J, Copeland TD, Waugh DS. The P1′ specificity of tobacco etch virus protease. Biochem Biophys Res Commun. 2002;294:949–55. doi: 10.1016/S0006-291X(02)00574-0. [DOI] [PubMed] [Google Scholar]
- 30.Panizzi P, Friedrich R, Fuentes-Prior P, Kroh HK, Briggs J, Tans G, Bode W, Bock PE. Novel fluorescent prothrombin analogs as probes of staphylocoagulase-prothrombin interactions. J Biol Chem. 2006;281:1169–78. doi: 10.1074/jbc.M507955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deutsch DG, Mertz ET. Plasminogen: purification from human plasma by affinity chromatography. Science. 1970;170:1095–6. doi: 10.1126/science.170.3962.1095. [DOI] [PubMed] [Google Scholar]
- 32.Castellino FJ, Powell JR. Human plasminogen. Methods Enzymol. 1981;80:365–78. doi: 10.1016/s0076-6879(81)80031-6. [DOI] [PubMed] [Google Scholar]
- 33.Bock PE. Active-site-selective labeling of blood coagulation proteinases with fluorescence probes by the use of thioester peptide chloromethyl ketones. I. Specificity of thrombin labeling. J Biol Chem. 1992;267:14963–73. [PubMed] [Google Scholar]
- 34.Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–47. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panizzi P, Swirski FK, Figueiredo JL, Waterman P, Sosnovik DE, Aikawa E, Libby P, Pittet M, Weissleder R, Nahrendorf M. Impaired infarct healing in atherosclerotic mice with Ly-6C(hi) monocytosis. J Am Coll Cardiol. 2010;55:1629–38. doi: 10.1016/j.jacc.2009.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, Figueiredo JL, Kohler RH, Chudnovskiy A, Waterman P, Aikawa E, Mempel TR, Libby P, Weissleder R, Pittet MJ. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–6. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi GY, Chang BI, Chen SM, Wu DH, Wu HL. Function of streptokinase fragments in plasminogen activation. Biochem J. 1994;304(Pt 1):235–41. doi: 10.1042/bj3040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brockway WJ, Castellino FJ. A characterization of native streptokinase and altered streptokinase isolated from a human plasminogen activator complex. Biochemistry. 1974;13:2063–70. doi: 10.1021/bi00707a010. [DOI] [PubMed] [Google Scholar]
- 39.Siefring GE, Jr, Castellino FJ. Interaction of streptokinase with plasminogen. Isolation and characterization of a streptokinase degradation product. J Biol Chem. 1976;251:3913–20. [PubMed] [Google Scholar]
- 40.Parrado J, Conejero-Lara F, Smith RA, Marshall JM, Ponting CP, Dobson CM. The domain organization of streptokinase: nuclear magnetic resonance, circular dichroism, and functional characterization of proteolytic fragments. Protein Sci. 1996;5:693–704. doi: 10.1002/pro.5560050414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajjar KA, Jacovina AT, Chacko J. An endothelial cell receptor for plasminogen/tissue plasminogen activator. I. Identity with annexin II. J Biol Chem. 1994;269:21191–7. [PubMed] [Google Scholar]
- 42.Cesarman GM, Guevara CA, Hajjar KA. An endothelial cell receptor for plasminogen/tissue plasminogen activator (t-PA). II. Annexin II-mediated enhancement of t-PA-dependent plasminogen activation. J Biol Chem. 1994;269:21198–203. [PubMed] [Google Scholar]
- 43.O’Connell PA, Surette AP, Liwski RS, Svenningsson P, Waisman DM. S100A10 regulates plasminogen-dependent macrophage invasion. Blood. 2010;116:1136–46. doi: 10.1182/blood-2010-01-264754. [DOI] [PubMed] [Google Scholar]
- 44.Andronicos NM, Chen EI, Baik N, Bai H, Parmer CM, Kiosses WB, Kamps MP, Yates JR, III, Parmer RJ, Miles LA. Proteomics-based discovery of a novel, structurally unique, and developmentally regulated plasminogen receptor, Plg-RKT, a major regulator of cell surface plasminogen activation. Blood. 2010;115:1319–1330. doi: 10.1182/blood-2008-11-188938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Das R, Burke T, Plow EF. Histone H2B as a functionally important plasminogen receptor on macrophages. Blood. 2007;110:3763–72. doi: 10.1182/blood-2007-03-079392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Gong Y, Grella DK, Castellino FJ, Miles LA. Endogenous plasmin converts Glu-plasminogen to Lys-plasminogen on the monocytoid cell surface. J Thromb Haemost. 2003;1:1264–70. doi: 10.1046/j.1538-7836.2003.00155.x. [DOI] [PubMed] [Google Scholar]
- 47.Miles LA, Castellino FJ, Gong Y. Critical role for conversion of glu-plasminogen to Lys-plasminogen for optimal stimulation of plasminogen activation on cell surfaces. Trends Cardiovasc Med. 2003;13:21–30. doi: 10.1016/s1050-1738(02)00190-1. [DOI] [PubMed] [Google Scholar]
- 48.Markus G, Evers JL, Hobika GH. Comparison of some properties of native (Glu) and modified (Lys) human plasminogen. J Biol Chem. 1978;253:733–9. [PubMed] [Google Scholar]
- 49.Marshall JM, Brown AJ, Ponting CP. Conformational studies of human plasminogen and plasminogen fragments: evidence for a novel third conformation of plasminogen. Biochemistry. 1994;33:3599–606. doi: 10.1021/bi00178a017. [DOI] [PubMed] [Google Scholar]
- 50.Violand BN, Byrne R, Castellino FJ. The effect of alpha-,omega-amino acids on human plasminogen structure and activation. J Biol Chem. 1978;253:5395–401. [PubMed] [Google Scholar]
- 51.Brownstein C, Deora AB, Jacovina AT, Weintraub R, Gertler M, Khan KM, Falcone DJ, Hajjar KA. Annexin II mediates plasminogen-dependent matrix invasion by human monocytes: enhanced expression by macrophages. Blood. 2004;103:317–24. doi: 10.1182/blood-2003-04-1304. [DOI] [PubMed] [Google Scholar]
- 52.Falcone DJ, Borth W, Khan KM, Hajjar KA. Plasminogen-mediated matrix invasion and degradation by macrophages is dependent on surface expression of annexin II. Blood. 2001;97:777–84. doi: 10.1182/blood.v97.3.777. [DOI] [PubMed] [Google Scholar]
- 53.Castellino FJ, Ploplis VA. Structure and function of the plasminogen/plasmin system. Thromb Haemost. 2005;93:647–54. doi: 10.1160/TH04-12-0842. [DOI] [PubMed] [Google Scholar]
- 54.Plow EF, Herren T, Redlitz A, Miles LA, Hoover-Plow JL. The cell biology of the plasminogen system. Faseb J. 1995;9:939–45. doi: 10.1096/fasebj.9.10.7615163. [DOI] [PubMed] [Google Scholar]
- 55.Laumonnier Y, Syrovets T, Burysek L, Simmet T. Identification of the annexin A2 heterotetramer as a receptor for the plasmin-induced signaling in human peripheral monocytes. Blood. 2006;107:3342–9. doi: 10.1182/blood-2005-07-2840. [DOI] [PubMed] [Google Scholar]
- 56.Andreasen PA, Kjoller L, Christensen L, Duffy MJ. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 57.Dass K, Ahmad A, Azmi AS, Sarkar SH, Sarkar FH. Evolving role of uPA/uPAR system in human cancers. Cancer Treat Rev. 2008;34:122–36. doi: 10.1016/j.ctrv.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Palumbo JS, Talmage KE, Liu H, La Jeunesse CM, Witte DP, Degen JL. Plasminogen supports tumor growth through a fibrinogen-dependent mechanism linked to vascular patency. Blood. 2003;102:2819–27. doi: 10.1182/blood-2003-03-0881. [DOI] [PubMed] [Google Scholar]
- 59.McMahon B, Kwaan HC. The Plasminogen Activator System and Cancer. Pathophysiology of Haemostasis and Thrombosis. 2007;36:184–194. doi: 10.1159/000175156. [DOI] [PubMed] [Google Scholar]
- 60.Semov A, Moreno MJ, Onichtchenko A, Abulrob A, Ball M, Ekiel I, Pietrzynski G, Stanimirovic D, Alakhov V. Metastasis-associated protein S100A4 induces angiogenesis through interaction with Annexin II and accelerated plasmin formation. J Biol Chem. 2005;280:20833–41. doi: 10.1074/jbc.M412653200. [DOI] [PubMed] [Google Scholar]
- 61.Ranson M, Andronicos NM, O’Mullane MJ, Baker MS. Increased plasminogen binding is associated with metastatic breast cancer cells: differential expression of plasminogen binding proteins. Br J Cancer. 1998;77:1586–97. doi: 10.1038/bjc.1998.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Law B, Curino A, Bugge TH, Weissleder R, Tung CH. Design, synthesis, and characterization of urokinase plasminogen-activator-sensitive near-infrared reporter. Chem Biol. 2004;11:99–106. doi: 10.1016/j.chembiol.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 63.Stillfried GE, Saunders DN, Ranson M. Plasminogen binding and activation at the breast cancer cell surface: the integral role of urokinase activity. Breast Cancer Res. 2007;9:R14. doi: 10.1186/bcr1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–62. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]