Fig. 4.
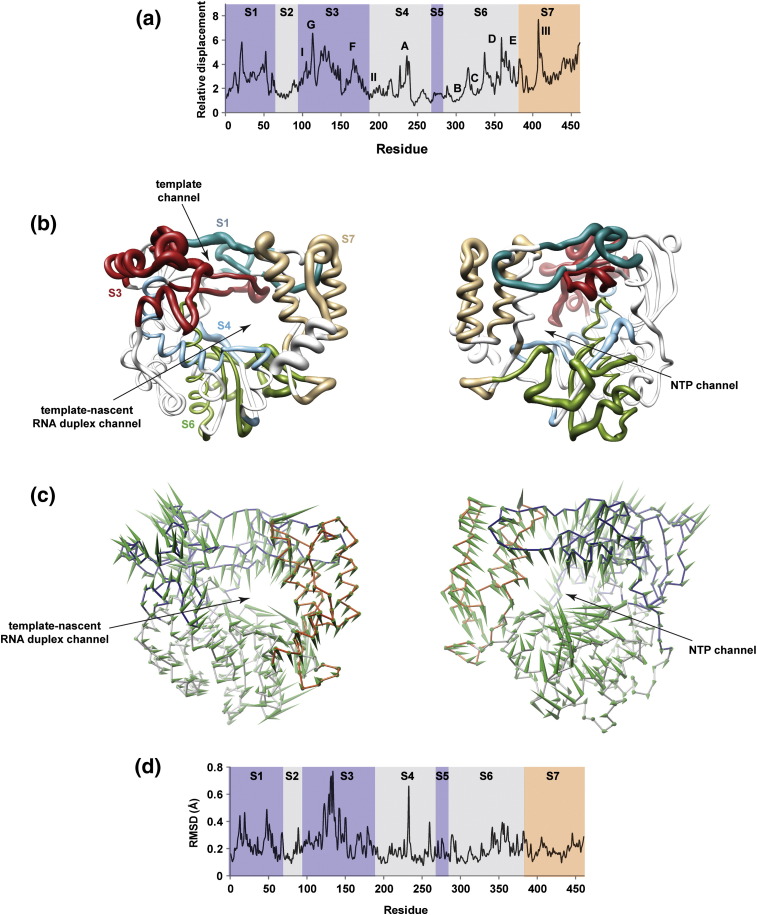
Functional motifs of PV RdRp largely contribute to the major motions observed during the simulation. (a) The relative displacements of Cα atoms observed in the sum of the first six PCs (modes) are plotted for each residue. Peaks correspond to regions contributing to the major motions observed during the MD simulations. The structure has been divided into seven segments (S1–S7). The fingers subdomain is shaded purple, the palm subdomain is shaded light gray and the thumb subdomain is shaded orange. The conserved motifs A-G are indicated. Other regions of PV RdRp known to have functional significance have been labeled I–III. (b) PV RdRp structure rendered as a tube to indicate flexibility and shown in two views: (left) looking through the template–nascent RNA duplex channel and (right) looking through the NTP channel. The radius of the tube indicates the magnitude of the relative displacement shown in (a). Each segment has a unique color. Only segments S1, S3, S4, S6 and S7 are shown. Interestingly, highly dynamic regions are located around the nascent RNA duplex and NTP channels. (c) The motion content of PC1, the largest mode of PCA, is visualized in two views that show the PV RdRp from the front (left) and the back (right). The starting structure of PV RdRp is depicted as wires with subdomains colored differently: blue (fingers), gray (palm) and red (thumb). The green “porcupine needles” indicate the direction of displacements of motions based on PC1; the size of the needle is proportional to the displacement. From the directions of displacement vectors represented by the needles, expansion and contraction of the RNA and NTP channels can be indicated; interestingly, when the template–nascent RNA duplex channel is open, the NTP channel is closed and vice versa. (d) Structural variation among multiple crystal structures of PV RdRp is represented by RMSD for each residue in PV RdRp, calculated from superimposed crystallographic structures (PDB codes 1RA6, 1RA7, 2IM0, 2IM1, 2IM2, 2IM3, 2ILY and 2ILZ). The structures were superimposed using Cα atoms of motifs B and C; superimposition was performed using Chimera. Residues that demonstrate large structural variations, indicated by high RMSD values, are implied to be flexible. Structural variations among different crystal structures qualitatively showed a pattern similar to that displayed in (a) for residues in segments S1–S5. The smaller amount of variability of residues in segment S6 could be a result of using residues within this segment for superimposition. Differences between experimental data and simulation observed for residues in segment S7 are attributable to the crystal packing effect where mobility of these residues is masked inside crystals; such an effect is absent from simulation.